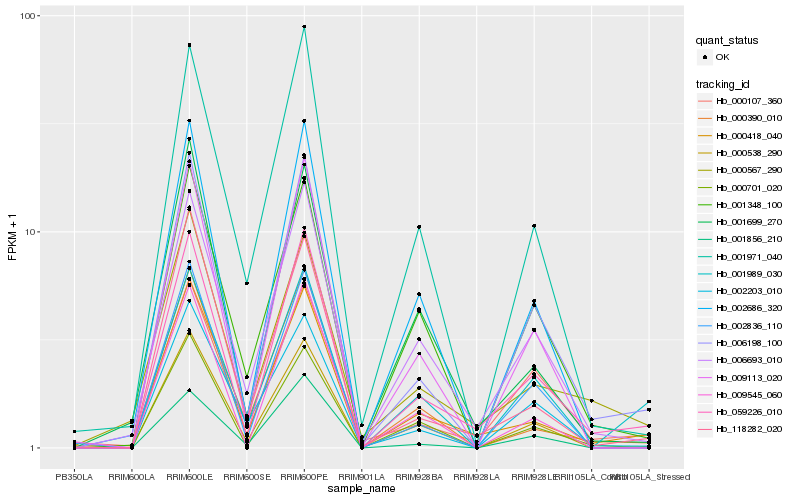
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_059226_010 |
0.0 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 2 |
Hb_006198_100 |
0.0862730519 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 3 |
Hb_000418_040 |
0.1002677341 |
- |
- |
hypothetical protein RCOM_0377590 [Ricinus communis] |
| 4 |
Hb_006693_010 |
0.1139837902 |
- |
- |
PREDICTED: uncharacterized protein LOC105628875 [Jatropha curcas] |
| 5 |
Hb_000538_290 |
0.1208558934 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 6 |
Hb_000107_360 |
0.1248478573 |
- |
- |
PREDICTED: xylem cysteine proteinase 1 [Jatropha curcas] |
| 7 |
Hb_009113_020 |
0.1282454096 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 8 |
Hb_000701_020 |
0.1351303471 |
- |
- |
PREDICTED: protein ENDOSPERM DEFECTIVE 1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000390_010 |
0.1376197848 |
- |
- |
PREDICTED: protein CDI [Jatropha curcas] |
| 10 |
Hb_118282_020 |
0.1376303942 |
- |
- |
PREDICTED: uncharacterized protein LOC105631532 isoform X3 [Jatropha curcas] |
| 11 |
Hb_002686_320 |
0.1377203471 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 12 |
Hb_001971_040 |
0.1391909642 |
- |
- |
PREDICTED: beta-glucosidase 44-like [Jatropha curcas] |
| 13 |
Hb_002836_110 |
0.1423357851 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like protein 8 [Jatropha curcas] |
| 14 |
Hb_000567_290 |
0.1441971234 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 15 |
Hb_002203_010 |
0.145302491 |
- |
- |
PREDICTED: kinesin-like protein KIF18B isoform X1 [Vitis vinifera] |
| 16 |
Hb_001989_030 |
0.1454456347 |
- |
- |
PREDICTED: copper transport protein ATX1 [Jatropha curcas] |
| 17 |
Hb_009545_060 |
0.1459744798 |
- |
- |
PREDICTED: uncharacterized protein LOC105632005 [Jatropha curcas] |
| 18 |
Hb_001348_100 |
0.1470217034 |
- |
- |
PREDICTED: uncharacterized protein LOC105638934 [Jatropha curcas] |
| 19 |
Hb_001856_210 |
0.1476092546 |
- |
- |
PREDICTED: uncharacterized protein LOC105644171 [Jatropha curcas] |
| 20 |
Hb_001699_270 |
0.1488571872 |
- |
- |
PREDICTED: uncharacterized protein LOC105628996 [Jatropha curcas] |