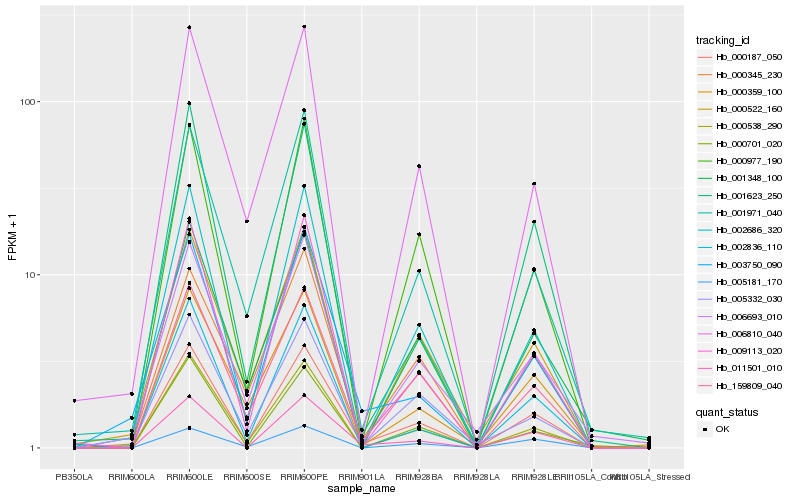
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000187_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650800 [Jatropha curcas] |
| 2 |
Hb_159809_040 |
0.0812102606 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002686_320 |
0.0878680179 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 4 |
Hb_011501_010 |
0.0879927815 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG5-like [Jatropha curcas] |
| 5 |
Hb_000538_290 |
0.09224411 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 6 |
Hb_000977_190 |
0.0947479347 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_009113_020 |
0.0959559435 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 8 |
Hb_001971_040 |
0.108037323 |
- |
- |
PREDICTED: beta-glucosidase 44-like [Jatropha curcas] |
| 9 |
Hb_005332_030 |
0.1090967739 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000359_100 |
0.1093239465 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_006810_040 |
0.1096800171 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 12 |
Hb_000345_230 |
0.1121789628 |
- |
- |
- |
| 13 |
Hb_001348_100 |
0.1136489003 |
- |
- |
PREDICTED: uncharacterized protein LOC105638934 [Jatropha curcas] |
| 14 |
Hb_006693_010 |
0.1153115473 |
- |
- |
PREDICTED: uncharacterized protein LOC105628875 [Jatropha curcas] |
| 15 |
Hb_002836_110 |
0.1162453518 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like protein 8 [Jatropha curcas] |
| 16 |
Hb_000701_020 |
0.116622667 |
- |
- |
PREDICTED: protein ENDOSPERM DEFECTIVE 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_005181_170 |
0.1172766144 |
- |
- |
PREDICTED: aldehyde dehydrogenase 22A1 [Jatropha curcas] |
| 18 |
Hb_003750_090 |
0.1172890792 |
- |
- |
BnaC02g34020D [Brassica napus] |
| 19 |
Hb_000522_160 |
0.1175056964 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 20 |
Hb_001623_250 |
0.1214011698 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |