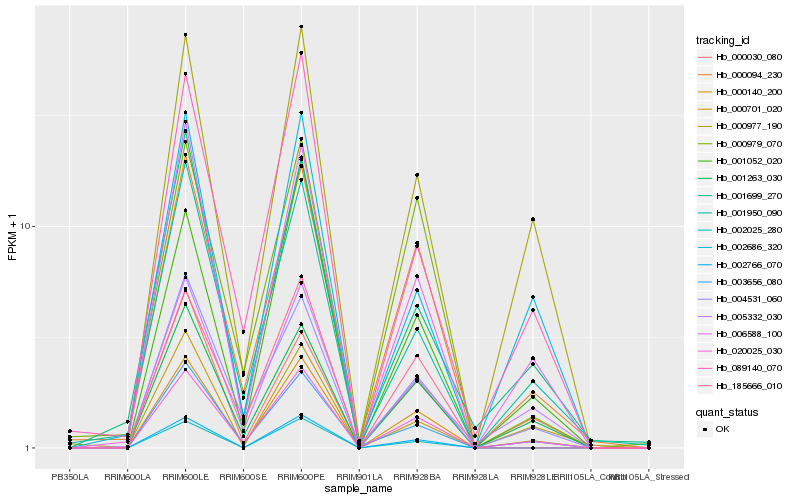
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000140_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642928 [Jatropha curcas] |
| 2 |
Hb_020025_030 |
0.0341719487 |
transcription factor |
TF Family: PHD |
PREDICTED: protein Jade-1 [Populus euphratica] |
| 3 |
Hb_006588_100 |
0.0817135723 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000094_230 |
0.0847832692 |
- |
- |
hypothetical protein JCGZ_04416 [Jatropha curcas] |
| 5 |
Hb_001263_030 |
0.0866611255 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 6 |
Hb_003656_080 |
0.0900943153 |
- |
- |
PREDICTED: corepressor interacting with RBPJ 1-like [Jatropha curcas] |
| 7 |
Hb_000977_190 |
0.0920539937 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004531_060 |
0.099491354 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3 [Jatropha curcas] |
| 9 |
Hb_005332_030 |
0.1031870834 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001052_020 |
0.1233955382 |
- |
- |
hypothetical protein JCGZ_26487 [Jatropha curcas] |
| 11 |
Hb_089140_070 |
0.1235233161 |
- |
- |
PREDICTED: uncharacterized protein LOC105637961 [Jatropha curcas] |
| 12 |
Hb_001950_090 |
0.124346213 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1C [Jatropha curcas] |
| 13 |
Hb_002766_070 |
0.1257653558 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_17894 [Jatropha curcas] |
| 14 |
Hb_000979_070 |
0.1265629405 |
- |
- |
PREDICTED: endoglucanase 8-like [Jatropha curcas] |
| 15 |
Hb_002686_320 |
0.1265877222 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 16 |
Hb_185666_010 |
0.1279832874 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3 [Jatropha curcas] |
| 17 |
Hb_000030_080 |
0.1284060048 |
- |
- |
PREDICTED: uncharacterized protein LOC105632640 [Jatropha curcas] |
| 18 |
Hb_000701_020 |
0.1309248968 |
- |
- |
PREDICTED: protein ENDOSPERM DEFECTIVE 1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_002025_280 |
0.1313790689 |
- |
- |
PREDICTED: uncharacterized protein LOC105634201 [Jatropha curcas] |
| 20 |
Hb_001699_270 |
0.1329447176 |
- |
- |
PREDICTED: uncharacterized protein LOC105628996 [Jatropha curcas] |