| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
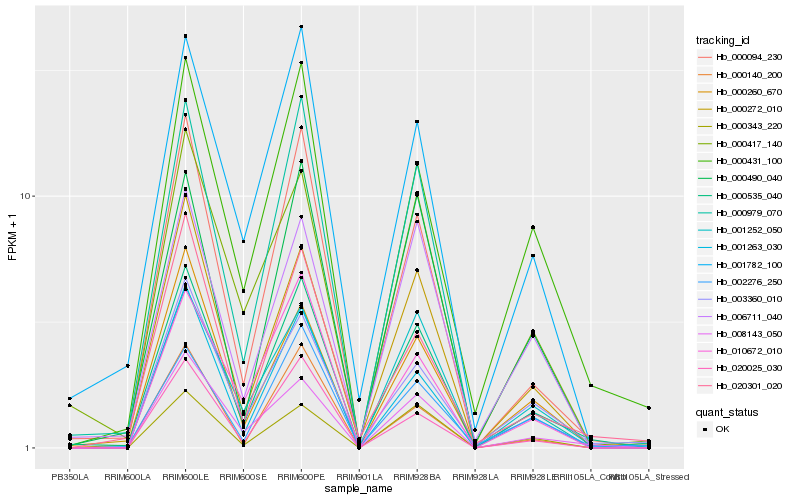
Hb_000535_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642104 [Jatropha curcas] |
| 2 |
Hb_000094_230 |
0.08001822 |
- |
- |
hypothetical protein JCGZ_04416 [Jatropha curcas] |
| 3 |
Hb_000272_010 |
0.101305502 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 4 |
Hb_008143_050 |
0.1034003003 |
- |
- |
PREDICTED: cytochrome P450 710A1-like [Jatropha curcas] |
| 5 |
Hb_001782_100 |
0.1137140794 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 6 |
Hb_001263_030 |
0.1140918519 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 7 |
Hb_000979_070 |
0.1170084415 |
- |
- |
PREDICTED: endoglucanase 8-like [Jatropha curcas] |
| 8 |
Hb_001252_050 |
0.1193758648 |
- |
- |
Wall-associated receptor kinase-like 20 [Morus notabilis] |
| 9 |
Hb_000431_100 |
0.1222452541 |
- |
- |
ribonucleoside-diphosphate reductase small chain, putative [Ricinus communis] |
| 10 |
Hb_000490_040 |
0.1230131647 |
- |
- |
PREDICTED: cyclin-D3-1 [Jatropha curcas] |
| 11 |
Hb_000260_670 |
0.123856418 |
- |
- |
PREDICTED: serine/threonine-protein kinase TIO isoform X1 [Jatropha curcas] |
| 12 |
Hb_000417_140 |
0.1243336522 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 2 [Jatropha curcas] |
| 13 |
Hb_006711_040 |
0.1267131845 |
- |
- |
PREDICTED: probable protein ABIL5 [Jatropha curcas] |
| 14 |
Hb_003360_010 |
0.1273246822 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 15 |
Hb_000343_220 |
0.1298911231 |
- |
- |
hypothetical protein CISIN_1g037489mg [Citrus sinensis] |
| 16 |
Hb_020301_020 |
0.1299072578 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 17 |
Hb_020025_030 |
0.1325643816 |
transcription factor |
TF Family: PHD |
PREDICTED: protein Jade-1 [Populus euphratica] |
| 18 |
Hb_002276_250 |
0.1333570784 |
- |
- |
PREDICTED: WD repeat and HMG-box DNA-binding protein 1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_010672_010 |
0.1367987509 |
- |
- |
hypothetical protein JCGZ_00895 [Jatropha curcas] |
| 20 |
Hb_000140_200 |
0.1390741931 |
- |
- |
PREDICTED: uncharacterized protein LOC105642928 [Jatropha curcas] |