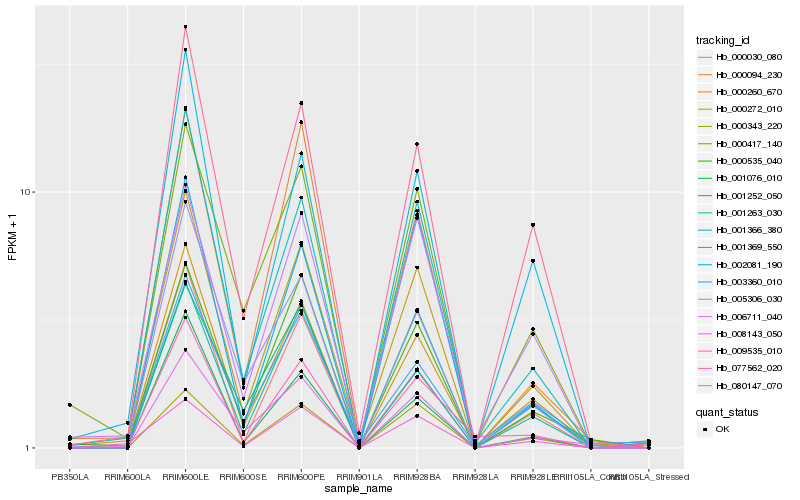
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000272_010 |
0.0 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 2 |
Hb_001366_380 |
0.0743351796 |
- |
- |
PREDICTED: pectin acetylesterase 6-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_000260_670 |
0.0771930766 |
- |
- |
PREDICTED: serine/threonine-protein kinase TIO isoform X1 [Jatropha curcas] |
| 4 |
Hb_080147_070 |
0.0948765653 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 5 |
Hb_000535_040 |
0.101305502 |
- |
- |
PREDICTED: uncharacterized protein LOC105642104 [Jatropha curcas] |
| 6 |
Hb_000343_220 |
0.1050352389 |
- |
- |
hypothetical protein CISIN_1g037489mg [Citrus sinensis] |
| 7 |
Hb_006711_040 |
0.1100176752 |
- |
- |
PREDICTED: probable protein ABIL5 [Jatropha curcas] |
| 8 |
Hb_001369_550 |
0.110632307 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 9 |
Hb_000417_140 |
0.1157740757 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 2 [Jatropha curcas] |
| 10 |
Hb_005306_030 |
0.1158727244 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 [Jatropha curcas] |
| 11 |
Hb_001263_030 |
0.1166747521 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 12 |
Hb_000094_230 |
0.1180393096 |
- |
- |
hypothetical protein JCGZ_04416 [Jatropha curcas] |
| 13 |
Hb_000030_080 |
0.1185020663 |
- |
- |
PREDICTED: uncharacterized protein LOC105632640 [Jatropha curcas] |
| 14 |
Hb_008143_050 |
0.1200455174 |
- |
- |
PREDICTED: cytochrome P450 710A1-like [Jatropha curcas] |
| 15 |
Hb_003360_010 |
0.1225002243 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 16 |
Hb_002081_190 |
0.1261736814 |
- |
- |
hypothetical protein EUGRSUZ_I010391, partial [Eucalyptus grandis] |
| 17 |
Hb_009535_010 |
0.1263883299 |
- |
- |
PREDICTED: DNA replication licensing factor MCM4 [Jatropha curcas] |
| 18 |
Hb_001076_010 |
0.1285114852 |
transcription factor |
TF Family: B3 |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_001252_050 |
0.1291159532 |
- |
- |
Wall-associated receptor kinase-like 20 [Morus notabilis] |
| 20 |
Hb_077562_020 |
0.1328669369 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 5 [Jatropha curcas] |