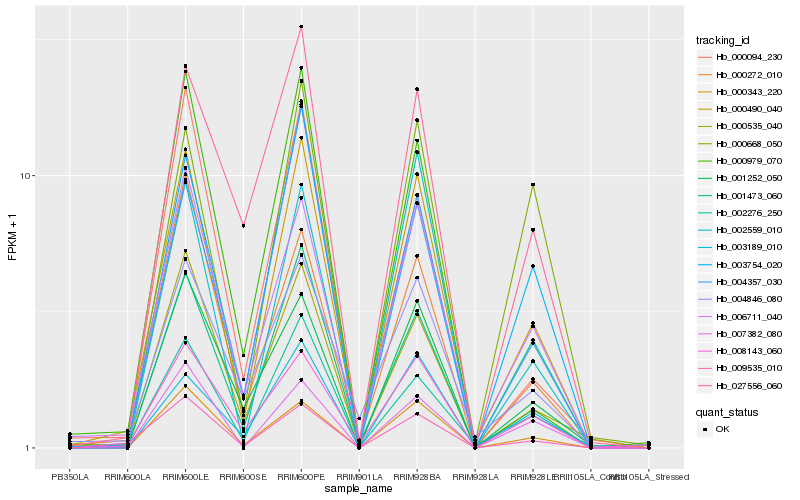
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000490_040 |
0.0 |
- |
- |
PREDICTED: cyclin-D3-1 [Jatropha curcas] |
| 2 |
Hb_006711_040 |
0.0975834501 |
- |
- |
PREDICTED: probable protein ABIL5 [Jatropha curcas] |
| 3 |
Hb_000343_220 |
0.1049804388 |
- |
- |
hypothetical protein CISIN_1g037489mg [Citrus sinensis] |
| 4 |
Hb_009535_010 |
0.1114589276 |
- |
- |
PREDICTED: DNA replication licensing factor MCM4 [Jatropha curcas] |
| 5 |
Hb_002276_250 |
0.1197281091 |
- |
- |
PREDICTED: WD repeat and HMG-box DNA-binding protein 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_004357_030 |
0.1212387791 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 7 |
Hb_000535_040 |
0.1230131647 |
- |
- |
PREDICTED: uncharacterized protein LOC105642104 [Jatropha curcas] |
| 8 |
Hb_003754_020 |
0.1294976421 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_008143_060 |
0.131613046 |
- |
- |
PREDICTED: putative hydrolase C777.06c isoform X2 [Jatropha curcas] |
| 10 |
Hb_001252_050 |
0.136202432 |
- |
- |
Wall-associated receptor kinase-like 20 [Morus notabilis] |
| 11 |
Hb_002559_010 |
0.1387275247 |
- |
- |
hypothetical protein JCGZ_06274 [Jatropha curcas] |
| 12 |
Hb_007382_080 |
0.1388652353 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 13 |
Hb_000272_010 |
0.1399164176 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 14 |
Hb_003189_010 |
0.1413520258 |
- |
- |
hypothetical protein EUGRSUZ_G02605 [Eucalyptus grandis] |
| 15 |
Hb_001473_060 |
0.1419566663 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 8 [Jatropha curcas] |
| 16 |
Hb_004846_080 |
0.1486632448 |
- |
- |
DNA replication licensing factor MCM3, putative [Ricinus communis] |
| 17 |
Hb_000668_050 |
0.1499983289 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 18 |
Hb_000979_070 |
0.1507485091 |
- |
- |
PREDICTED: endoglucanase 8-like [Jatropha curcas] |
| 19 |
Hb_027556_060 |
0.1507834231 |
- |
- |
hypothetical protein POPTR_0001s03520g [Populus trichocarpa] |
| 20 |
Hb_000094_230 |
0.1547035346 |
- |
- |
hypothetical protein JCGZ_04416 [Jatropha curcas] |