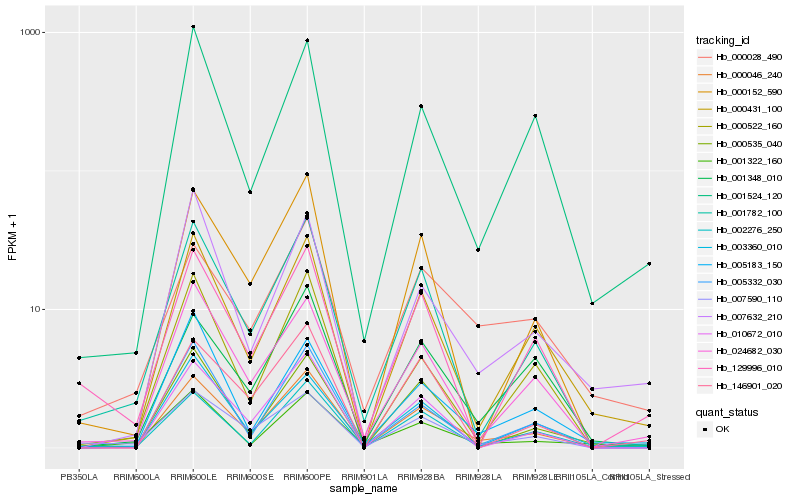
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_240 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631837 [Jatropha curcas] |
| 2 |
Hb_000431_100 |
0.1117210824 |
- |
- |
ribonucleoside-diphosphate reductase small chain, putative [Ricinus communis] |
| 3 |
Hb_002276_250 |
0.1206637497 |
- |
- |
PREDICTED: WD repeat and HMG-box DNA-binding protein 1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_024682_030 |
0.1222824518 |
- |
- |
PREDICTED: DNA replication licensing factor MCM2 [Jatropha curcas] |
| 5 |
Hb_001782_100 |
0.1264789461 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 6 |
Hb_001524_120 |
0.1316891004 |
- |
- |
myo-inositol-1 phosphate synthase [Hevea brasiliensis] |
| 7 |
Hb_010672_010 |
0.1405705332 |
- |
- |
hypothetical protein JCGZ_00895 [Jatropha curcas] |
| 8 |
Hb_000152_590 |
0.1413356113 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 9 |
Hb_003360_010 |
0.1421973537 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 10 |
Hb_001348_010 |
0.1442280223 |
- |
- |
PREDICTED: lysM domain-containing GPI-anchored protein 1 [Jatropha curcas] |
| 11 |
Hb_007632_210 |
0.1447556989 |
- |
- |
PREDICTED: sodium/potassium/calcium exchanger 1 [Populus euphratica] |
| 12 |
Hb_005332_030 |
0.1485943085 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_146901_020 |
0.1513720427 |
- |
- |
- |
| 14 |
Hb_005183_150 |
0.1527132076 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 15 |
Hb_000535_040 |
0.1537512696 |
- |
- |
PREDICTED: uncharacterized protein LOC105642104 [Jatropha curcas] |
| 16 |
Hb_000522_160 |
0.1547556957 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 17 |
Hb_001322_160 |
0.1548758021 |
- |
- |
hypothetical protein JCGZ_03472 [Jatropha curcas] |
| 18 |
Hb_007590_110 |
0.1557973553 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |
| 19 |
Hb_129996_010 |
0.1572968835 |
- |
- |
hypothetical protein PRUPE_ppa013547mg [Prunus persica] |
| 20 |
Hb_000028_490 |
0.1579635602 |
- |
- |
calmodulin-like protein 6a [Populus trichocarpa] |