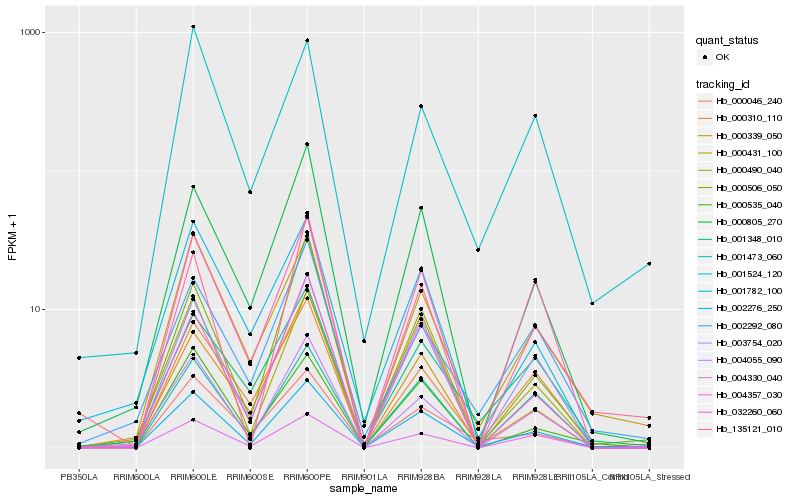
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002276_250 |
0.0 |
- |
- |
PREDICTED: WD repeat and HMG-box DNA-binding protein 1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_003754_020 |
0.1021806891 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_000431_100 |
0.1037756028 |
- |
- |
ribonucleoside-diphosphate reductase small chain, putative [Ricinus communis] |
| 4 |
Hb_004055_090 |
0.106878526 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 [Jatropha curcas] |
| 5 |
Hb_000805_270 |
0.1157811628 |
- |
- |
PREDICTED: expansin-A4 [Gossypium raimondii] |
| 6 |
Hb_000490_040 |
0.1197281091 |
- |
- |
PREDICTED: cyclin-D3-1 [Jatropha curcas] |
| 7 |
Hb_000046_240 |
0.1206637497 |
- |
- |
PREDICTED: uncharacterized protein LOC105631837 [Jatropha curcas] |
| 8 |
Hb_004357_030 |
0.1221383322 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 9 |
Hb_001473_060 |
0.1307639999 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 8 [Jatropha curcas] |
| 10 |
Hb_004330_040 |
0.1317797439 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001348_010 |
0.1330760473 |
- |
- |
PREDICTED: lysM domain-containing GPI-anchored protein 1 [Jatropha curcas] |
| 12 |
Hb_000535_040 |
0.1333570784 |
- |
- |
PREDICTED: uncharacterized protein LOC105642104 [Jatropha curcas] |
| 13 |
Hb_000339_050 |
0.13635143 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 2 [Jatropha curcas] |
| 14 |
Hb_000506_050 |
0.1398131466 |
- |
- |
PREDICTED: thymidine kinase [Jatropha curcas] |
| 15 |
Hb_002292_080 |
0.1403206255 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 16 |
Hb_000310_110 |
0.1403288524 |
- |
- |
PREDICTED: probable trehalose-phosphate phosphatase F isoform X1 [Jatropha curcas] |
| 17 |
Hb_001782_100 |
0.1405963645 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 18 |
Hb_001524_120 |
0.1406524621 |
- |
- |
myo-inositol-1 phosphate synthase [Hevea brasiliensis] |
| 19 |
Hb_032260_060 |
0.1414716383 |
transcription factor |
TF Family: MYB-related |
Homeodomain-like superfamily protein [Theobroma cacao] |
| 20 |
Hb_135121_010 |
0.1451364408 |
- |
- |
PREDICTED: uncharacterized protein LOC105635544 [Jatropha curcas] |