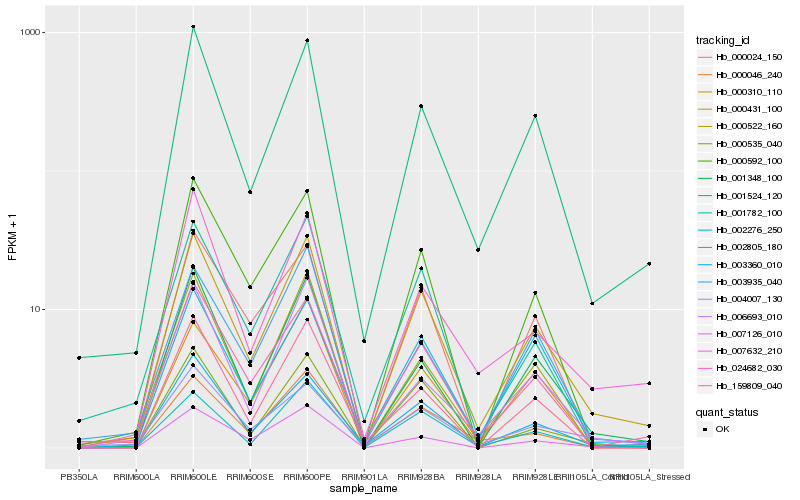
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000431_100 |
0.0 |
- |
- |
ribonucleoside-diphosphate reductase small chain, putative [Ricinus communis] |
| 2 |
Hb_001524_120 |
0.0801752335 |
- |
- |
myo-inositol-1 phosphate synthase [Hevea brasiliensis] |
| 3 |
Hb_003360_010 |
0.0891994846 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 4 |
Hb_024682_030 |
0.0965568936 |
- |
- |
PREDICTED: DNA replication licensing factor MCM2 [Jatropha curcas] |
| 5 |
Hb_002276_250 |
0.1037756028 |
- |
- |
PREDICTED: WD repeat and HMG-box DNA-binding protein 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001348_100 |
0.1108579111 |
- |
- |
PREDICTED: uncharacterized protein LOC105638934 [Jatropha curcas] |
| 7 |
Hb_000046_240 |
0.1117210824 |
- |
- |
PREDICTED: uncharacterized protein LOC105631837 [Jatropha curcas] |
| 8 |
Hb_000592_100 |
0.1135565162 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 9 |
Hb_000522_160 |
0.1163875659 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 10 |
Hb_000535_040 |
0.1222452541 |
- |
- |
PREDICTED: uncharacterized protein LOC105642104 [Jatropha curcas] |
| 11 |
Hb_002805_180 |
0.1245982708 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004007_130 |
0.1253171866 |
- |
- |
PREDICTED: uncharacterized protein LOC105639843 [Jatropha curcas] |
| 13 |
Hb_007126_010 |
0.1261233277 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 14 |
Hb_001782_100 |
0.1277829998 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 15 |
Hb_000024_150 |
0.1285716293 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 16 |
Hb_003935_040 |
0.1296833443 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein SVP [Jatropha curcas] |
| 17 |
Hb_000310_110 |
0.1304110818 |
- |
- |
PREDICTED: probable trehalose-phosphate phosphatase F isoform X1 [Jatropha curcas] |
| 18 |
Hb_159809_040 |
0.1310334589 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_006693_010 |
0.1311790368 |
- |
- |
PREDICTED: uncharacterized protein LOC105628875 [Jatropha curcas] |
| 20 |
Hb_007632_210 |
0.1311999299 |
- |
- |
PREDICTED: sodium/potassium/calcium exchanger 1 [Populus euphratica] |