| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
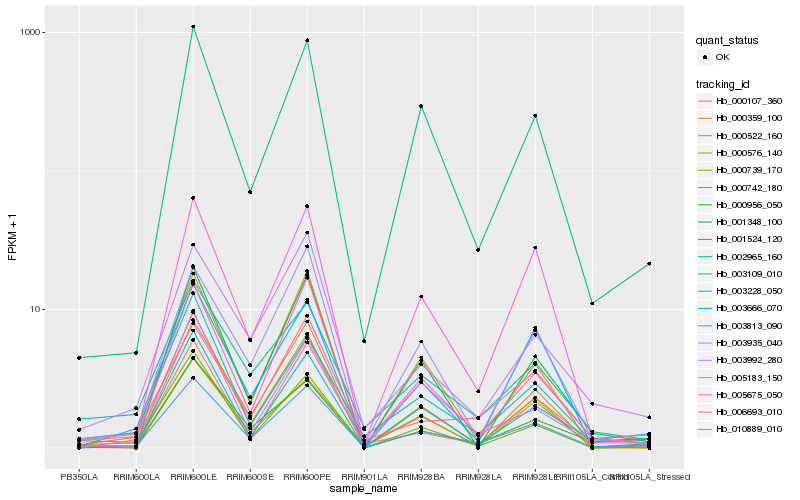
Hb_003813_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 2 |
Hb_001524_120 |
0.0923040002 |
- |
- |
myo-inositol-1 phosphate synthase [Hevea brasiliensis] |
| 3 |
Hb_000576_140 |
0.1146190143 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 4 |
Hb_006693_010 |
0.116569636 |
- |
- |
PREDICTED: uncharacterized protein LOC105628875 [Jatropha curcas] |
| 5 |
Hb_000956_050 |
0.116719009 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_003228_050 |
0.1170865581 |
- |
- |
chromomethylase [Hevea brasiliensis] |
| 7 |
Hb_005675_050 |
0.1209418123 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |
| 8 |
Hb_000522_160 |
0.1219119904 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 9 |
Hb_000107_360 |
0.1252999623 |
- |
- |
PREDICTED: xylem cysteine proteinase 1 [Jatropha curcas] |
| 10 |
Hb_010889_010 |
0.1291917192 |
- |
- |
PREDICTED: myosin-12 [Jatropha curcas] |
| 11 |
Hb_001348_100 |
0.1306092139 |
- |
- |
PREDICTED: uncharacterized protein LOC105638934 [Jatropha curcas] |
| 12 |
Hb_000742_180 |
0.1310710912 |
- |
- |
hypothetical protein POPTR_0011s16640g [Populus trichocarpa] |
| 13 |
Hb_002965_160 |
0.134128922 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |
| 14 |
Hb_003109_010 |
0.1349485809 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 15 |
Hb_000739_170 |
0.135480139 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 16 |
Hb_000359_100 |
0.1362787185 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003666_070 |
0.1373315818 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g66560 [Jatropha curcas] |
| 18 |
Hb_003935_040 |
0.1376892624 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein SVP [Jatropha curcas] |
| 19 |
Hb_003992_280 |
0.1380693393 |
- |
- |
PREDICTED: uncharacterized protein LOC105117987 [Populus euphratica] |
| 20 |
Hb_005183_150 |
0.1394127606 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |