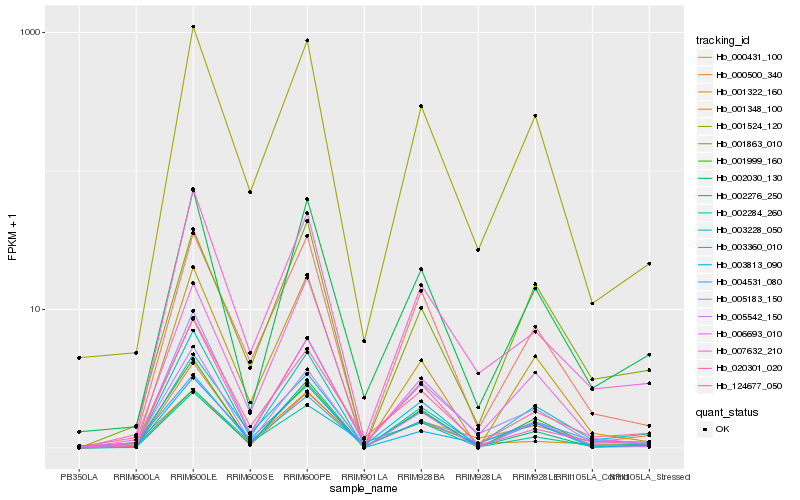
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002030_130 |
0.0 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 2 |
Hb_003228_050 |
0.1044218538 |
- |
- |
chromomethylase [Hevea brasiliensis] |
| 3 |
Hb_001524_120 |
0.1100861545 |
- |
- |
myo-inositol-1 phosphate synthase [Hevea brasiliensis] |
| 4 |
Hb_005183_150 |
0.1248468154 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 5 |
Hb_124677_050 |
0.1271701707 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_007632_210 |
0.1357224243 |
- |
- |
PREDICTED: sodium/potassium/calcium exchanger 1 [Populus euphratica] |
| 7 |
Hb_001999_160 |
0.1367935158 |
- |
- |
hypothetical protein JCGZ_08488 [Jatropha curcas] |
| 8 |
Hb_003360_010 |
0.1368086156 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 9 |
Hb_000431_100 |
0.1379291549 |
- |
- |
ribonucleoside-diphosphate reductase small chain, putative [Ricinus communis] |
| 10 |
Hb_002284_260 |
0.138326656 |
- |
- |
brca1 interacting protein helicase 1 brip1, putative [Ricinus communis] |
| 11 |
Hb_004531_080 |
0.1394293194 |
- |
- |
hypothetical protein RCOM_1491960 [Ricinus communis] |
| 12 |
Hb_005542_150 |
0.1401578753 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 13 |
Hb_001863_010 |
0.1438812356 |
- |
- |
PREDICTED: classical arabinogalactan protein 4-like [Jatropha curcas] |
| 14 |
Hb_003813_090 |
0.1470820242 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 15 |
Hb_020301_020 |
0.1479788871 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 16 |
Hb_000500_340 |
0.1482495107 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 17 |
Hb_002276_250 |
0.1488447771 |
- |
- |
PREDICTED: WD repeat and HMG-box DNA-binding protein 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001348_100 |
0.1498228425 |
- |
- |
PREDICTED: uncharacterized protein LOC105638934 [Jatropha curcas] |
| 19 |
Hb_006693_010 |
0.1509718013 |
- |
- |
PREDICTED: uncharacterized protein LOC105628875 [Jatropha curcas] |
| 20 |
Hb_001322_160 |
0.1534118619 |
- |
- |
hypothetical protein JCGZ_03472 [Jatropha curcas] |