| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
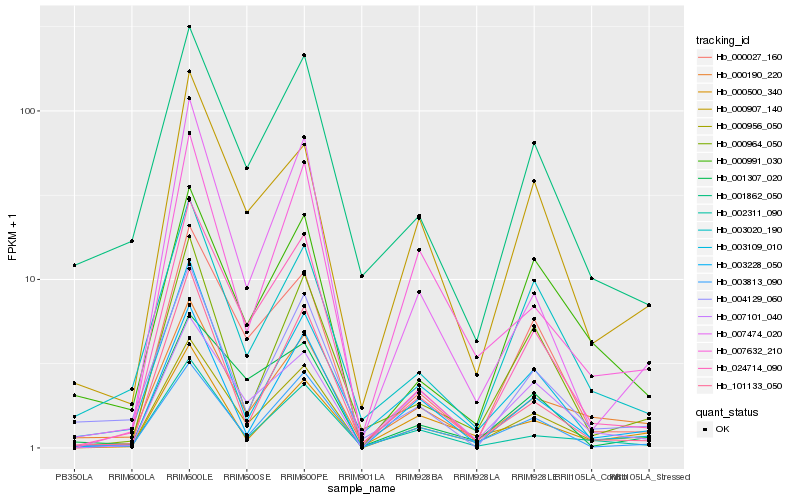
Hb_000956_050 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000190_220 |
0.1024621125 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek2 [Jatropha curcas] |
| 3 |
Hb_003228_050 |
0.1130658288 |
- |
- |
chromomethylase [Hevea brasiliensis] |
| 4 |
Hb_003813_090 |
0.116719009 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 5 |
Hb_024714_090 |
0.120959214 |
rubber biosynthesis |
Gene Name: SRPP3 |
PREDICTED: stress-related protein [Jatropha curcas] |
| 6 |
Hb_000907_140 |
0.1227391592 |
- |
- |
PREDICTED: annexin D5-like [Jatropha curcas] |
| 7 |
Hb_002311_090 |
0.1265882855 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |
| 8 |
Hb_000027_160 |
0.1285963602 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 9 |
Hb_007101_040 |
0.1399659808 |
- |
- |
PREDICTED: MLP-like protein 423 [Populus euphratica] |
| 10 |
Hb_001862_050 |
0.141349405 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 18-like [Jatropha curcas] |
| 11 |
Hb_000964_050 |
0.1425369476 |
- |
- |
PREDICTED: uncharacterized protein LOC105628694 [Jatropha curcas] |
| 12 |
Hb_001307_020 |
0.1428102818 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 13 |
Hb_007632_210 |
0.1430149342 |
- |
- |
PREDICTED: sodium/potassium/calcium exchanger 1 [Populus euphratica] |
| 14 |
Hb_101133_050 |
0.1438738696 |
- |
- |
PREDICTED: condensin complex subunit 3 [Jatropha curcas] |
| 15 |
Hb_000500_340 |
0.1439182826 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 16 |
Hb_003020_190 |
0.1444330382 |
- |
- |
PREDICTED: methylsterol monooxygenase 2-2 [Jatropha curcas] |
| 17 |
Hb_007474_020 |
0.1446829352 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 18 |
Hb_003109_010 |
0.1453349891 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 19 |
Hb_000991_030 |
0.1456956453 |
- |
- |
PREDICTED: uncharacterized protein LOC105122436 [Populus euphratica] |
| 20 |
Hb_004129_060 |
0.1458149987 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |