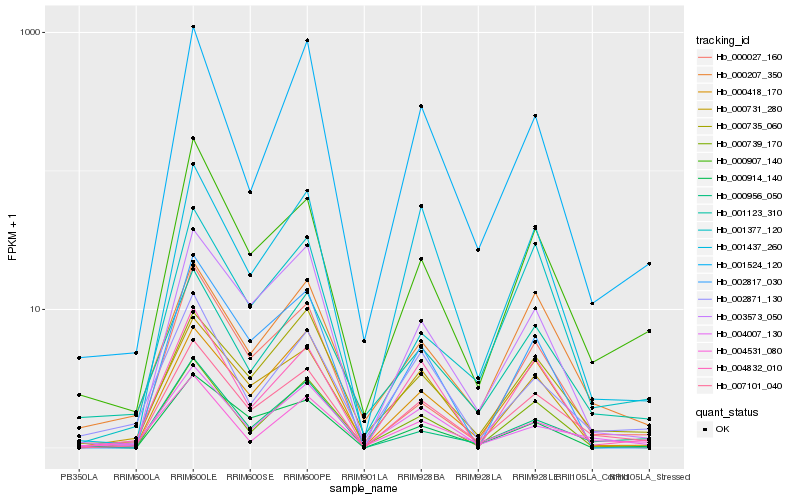
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007101_040 |
0.0 |
- |
- |
PREDICTED: MLP-like protein 423 [Populus euphratica] |
| 2 |
Hb_000907_140 |
0.1162620845 |
- |
- |
PREDICTED: annexin D5-like [Jatropha curcas] |
| 3 |
Hb_003573_050 |
0.1292521947 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 4 |
Hb_001437_260 |
0.1319214795 |
- |
- |
PREDICTED: protein FAM98B-like [Jatropha curcas] |
| 5 |
Hb_004007_130 |
0.1378659272 |
- |
- |
PREDICTED: uncharacterized protein LOC105639843 [Jatropha curcas] |
| 6 |
Hb_000914_140 |
0.1397446738 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_000956_050 |
0.1399659808 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_004832_010 |
0.140870884 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000735_060 |
0.1412038109 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 10 |
Hb_000731_280 |
0.1439161992 |
- |
- |
PREDICTED: uncharacterized protein LOC105649351 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000418_170 |
0.1443551584 |
- |
- |
PREDICTED: ABC transporter C family member 12-like isoform X4 [Populus euphratica] |
| 12 |
Hb_000207_350 |
0.1451888725 |
- |
- |
PREDICTED: embryogenesis-associated protein EMB8 [Jatropha curcas] |
| 13 |
Hb_001123_310 |
0.1452023587 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 14 |
Hb_000739_170 |
0.1462677022 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 15 |
Hb_001377_120 |
0.1486014411 |
- |
- |
PREDICTED: uncharacterized protein LOC105642861 [Jatropha curcas] |
| 16 |
Hb_001524_120 |
0.1499967508 |
- |
- |
myo-inositol-1 phosphate synthase [Hevea brasiliensis] |
| 17 |
Hb_002871_130 |
0.1501575844 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 4 homolog A isoform X2 [Jatropha curcas] |
| 18 |
Hb_004531_080 |
0.1502191954 |
- |
- |
hypothetical protein RCOM_1491960 [Ricinus communis] |
| 19 |
Hb_002817_030 |
0.1557140322 |
- |
- |
hypothetical protein POPTR_0001s39195g [Populus trichocarpa] |
| 20 |
Hb_000027_160 |
0.1558052979 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |