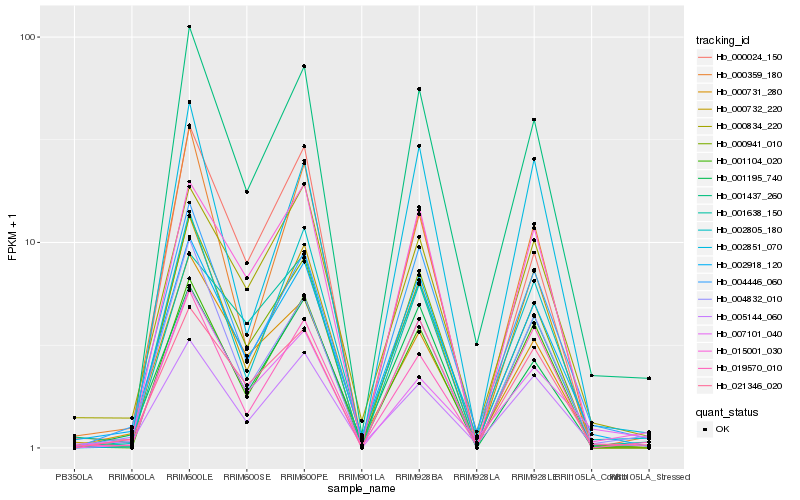
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001437_260 |
0.0 |
- |
- |
PREDICTED: protein FAM98B-like [Jatropha curcas] |
| 2 |
Hb_000941_010 |
0.070397562 |
- |
- |
hypothetical protein RCOM_0504460 [Ricinus communis] |
| 3 |
Hb_004446_060 |
0.0894786753 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 4 |
Hb_002805_180 |
0.093092922 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002918_120 |
0.0960704374 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_004832_010 |
0.1107528549 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001104_020 |
0.113457687 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_005144_060 |
0.1166716316 |
- |
- |
Disease resistance family protein / LRR family protein, putative [Theobroma cacao] |
| 9 |
Hb_000359_180 |
0.117593005 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000732_220 |
0.1182429943 |
- |
- |
PREDICTED: importin subunit alpha-5 [Jatropha curcas] |
| 11 |
Hb_015001_030 |
0.1195197296 |
- |
- |
inosine-uridine preferring nucleoside hydrolase family protein [Populus trichocarpa] |
| 12 |
Hb_021346_020 |
0.1204501294 |
- |
- |
hypothetical protein JCGZ_26746 [Jatropha curcas] |
| 13 |
Hb_000731_280 |
0.122726552 |
- |
- |
PREDICTED: uncharacterized protein LOC105649351 isoform X1 [Jatropha curcas] |
| 14 |
Hb_019570_010 |
0.1275392759 |
- |
- |
lyase, putative [Ricinus communis] |
| 15 |
Hb_000834_220 |
0.1297653228 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 16 |
Hb_001195_740 |
0.1299334265 |
transcription factor |
TF Family: LIM |
PREDICTED: protein DA1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000024_150 |
0.1311737869 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 18 |
Hb_002851_070 |
0.131384699 |
- |
- |
PREDICTED: uncharacterized protein LOC105646410 [Jatropha curcas] |
| 19 |
Hb_007101_040 |
0.1319214795 |
- |
- |
PREDICTED: MLP-like protein 423 [Populus euphratica] |
| 20 |
Hb_001638_150 |
0.1339332672 |
- |
- |
acyltransferase, putative [Ricinus communis] |