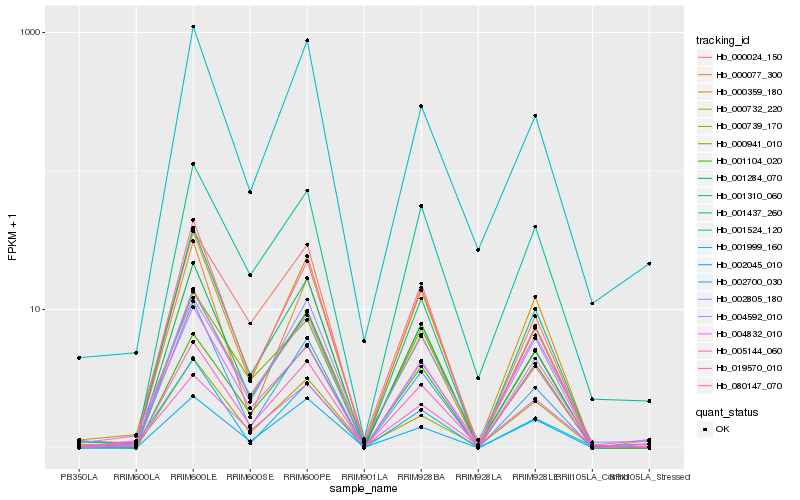
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000359_180 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002805_180 |
0.0806393548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000732_220 |
0.0908304179 |
- |
- |
PREDICTED: importin subunit alpha-5 [Jatropha curcas] |
| 4 |
Hb_001310_060 |
0.0914826473 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 5 |
Hb_000077_300 |
0.0991070658 |
- |
- |
PREDICTED: expansin-B3-like [Jatropha curcas] |
| 6 |
Hb_004832_010 |
0.0999176175 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004592_010 |
0.1006225508 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b-like [Jatropha curcas] |
| 8 |
Hb_002700_030 |
0.1039735325 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 9 |
Hb_019570_010 |
0.1040833752 |
- |
- |
lyase, putative [Ricinus communis] |
| 10 |
Hb_002045_010 |
0.1041918003 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 17 [Jatropha curcas] |
| 11 |
Hb_000739_170 |
0.1065827665 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 12 |
Hb_001104_020 |
0.1072431129 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_080147_070 |
0.1082126359 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 14 |
Hb_001284_070 |
0.1083312004 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000941_010 |
0.1104848807 |
- |
- |
hypothetical protein RCOM_0504460 [Ricinus communis] |
| 16 |
Hb_001524_120 |
0.1165234823 |
- |
- |
myo-inositol-1 phosphate synthase [Hevea brasiliensis] |
| 17 |
Hb_001437_260 |
0.117593005 |
- |
- |
PREDICTED: protein FAM98B-like [Jatropha curcas] |
| 18 |
Hb_001999_160 |
0.1208185818 |
- |
- |
hypothetical protein JCGZ_08488 [Jatropha curcas] |
| 19 |
Hb_000024_150 |
0.1280083664 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 20 |
Hb_005144_060 |
0.1293989461 |
- |
- |
Disease resistance family protein / LRR family protein, putative [Theobroma cacao] |