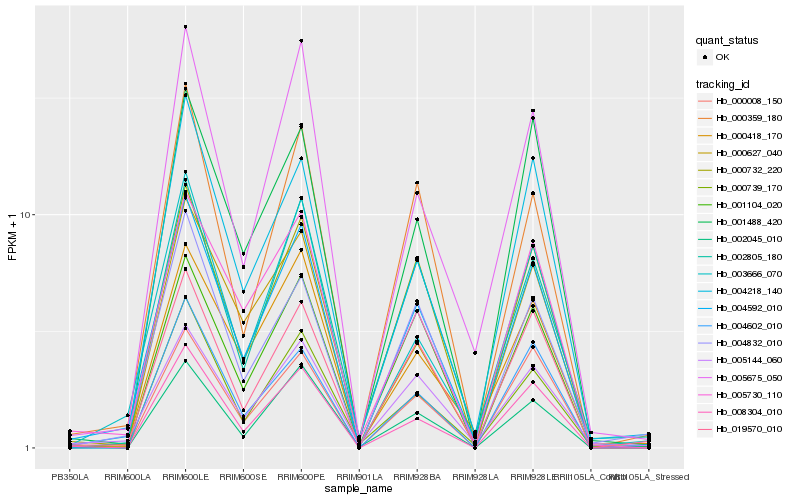
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004592_010 |
0.0 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b-like [Jatropha curcas] |
| 2 |
Hb_019570_010 |
0.0865423658 |
- |
- |
lyase, putative [Ricinus communis] |
| 3 |
Hb_003666_070 |
0.0924305465 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g66560 [Jatropha curcas] |
| 4 |
Hb_005730_110 |
0.092582965 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 5 |
Hb_008304_010 |
0.0928675435 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 6 |
Hb_000732_220 |
0.0996674127 |
- |
- |
PREDICTED: importin subunit alpha-5 [Jatropha curcas] |
| 7 |
Hb_000359_180 |
0.1006225508 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005144_060 |
0.1026506024 |
- |
- |
Disease resistance family protein / LRR family protein, putative [Theobroma cacao] |
| 9 |
Hb_000418_170 |
0.1031750886 |
- |
- |
PREDICTED: ABC transporter C family member 12-like isoform X4 [Populus euphratica] |
| 10 |
Hb_000739_170 |
0.1054100442 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 11 |
Hb_004218_140 |
0.1055612331 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 isoform X3 [Jatropha curcas] |
| 12 |
Hb_002805_180 |
0.1065374057 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005675_050 |
0.1076875279 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |
| 14 |
Hb_000008_150 |
0.1084598632 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 15 |
Hb_000627_040 |
0.1100297129 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 16 |
Hb_004832_010 |
0.111386904 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001104_020 |
0.1120406707 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_002045_010 |
0.113079487 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 17 [Jatropha curcas] |
| 19 |
Hb_001488_420 |
0.1133001585 |
- |
- |
PREDICTED: ABC transporter B family member 27 [Jatropha curcas] |
| 20 |
Hb_004602_010 |
0.1190833995 |
- |
- |
PREDICTED: uncharacterized protein LOC105042236 isoform X2 [Elaeis guineensis] |