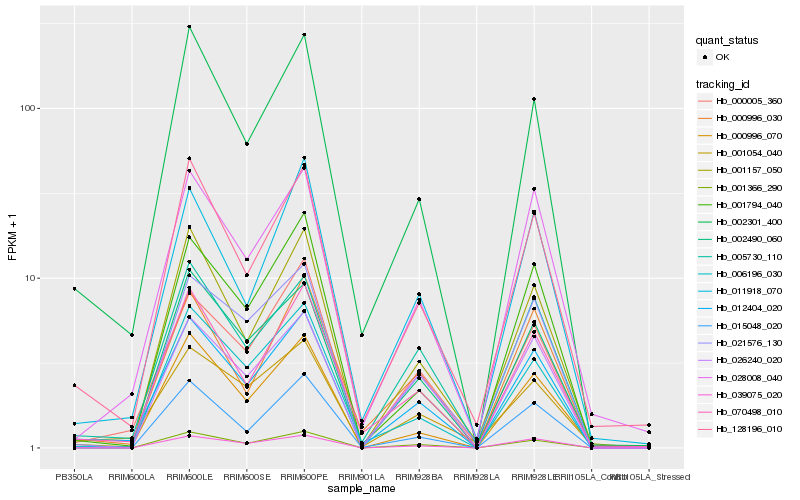
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012404_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_070498_010 |
0.0808723966 |
- |
- |
receptor kinase, putative [Ricinus communis] |
| 3 |
Hb_002301_400 |
0.0817585522 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 4 |
Hb_128196_010 |
0.0867007627 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005730_110 |
0.087332262 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 6 |
Hb_001366_290 |
0.0959291986 |
- |
- |
Alpha-expansin 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_028008_040 |
0.0972657614 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000005_360 |
0.0972741774 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_011918_070 |
0.1007215985 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 10 |
Hb_002490_060 |
0.1030820131 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 11 |
Hb_001157_050 |
0.1045139476 |
- |
- |
cation efflux protein/ zinc transporter, putative [Ricinus communis] |
| 12 |
Hb_039075_020 |
0.1079849731 |
- |
- |
PREDICTED: uncharacterized protein LOC103331245 [Prunus mume] |
| 13 |
Hb_006196_030 |
0.1080933462 |
- |
- |
PREDICTED: uncharacterized protein LOC105650679 [Jatropha curcas] |
| 14 |
Hb_001054_040 |
0.109706996 |
- |
- |
hypothetical protein JCGZ_25310 [Jatropha curcas] |
| 15 |
Hb_026240_020 |
0.1101148126 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 16 |
Hb_000996_030 |
0.110333573 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g63430 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000996_070 |
0.1105659135 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_021576_130 |
0.1109264583 |
- |
- |
PREDICTED: chorismate mutase 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001794_040 |
0.1112142569 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 20 |
Hb_015048_020 |
0.1119995939 |
- |
- |
PREDICTED: myb-like protein D [Jatropha curcas] |