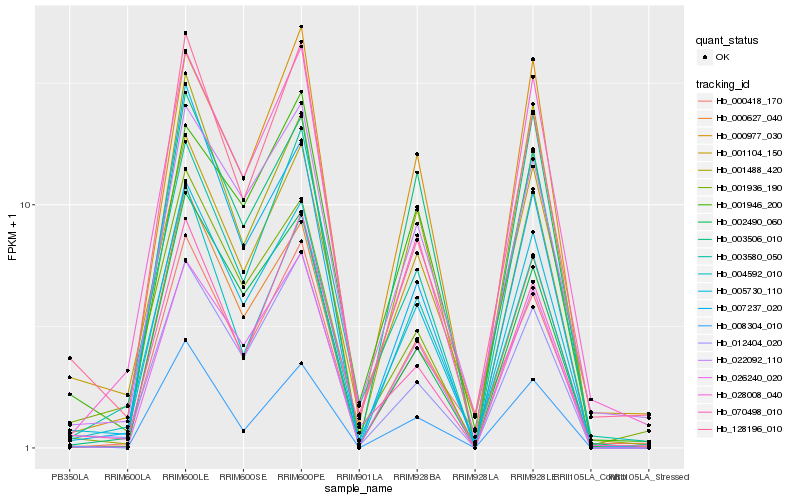
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005730_110 |
0.0 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 2 |
Hb_000627_040 |
0.0758374509 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 3 |
Hb_026240_020 |
0.0860207409 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 4 |
Hb_012404_020 |
0.087332262 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000418_170 |
0.0909906642 |
- |
- |
PREDICTED: ABC transporter C family member 12-like isoform X4 [Populus euphratica] |
| 6 |
Hb_022092_110 |
0.0919078382 |
- |
- |
PREDICTED: probable protein phosphatase 2C 52 [Jatropha curcas] |
| 7 |
Hb_004592_010 |
0.092582965 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b-like [Jatropha curcas] |
| 8 |
Hb_001488_420 |
0.093271061 |
- |
- |
PREDICTED: ABC transporter B family member 27 [Jatropha curcas] |
| 9 |
Hb_002490_060 |
0.0999169717 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 10 |
Hb_028008_040 |
0.1019350293 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000977_030 |
0.1022418135 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_128196_010 |
0.1033714451 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001104_150 |
0.1054588424 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 14 |
Hb_003580_050 |
0.1077192143 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 15 |
Hb_001946_200 |
0.1083452549 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 16 |
Hb_003506_010 |
0.1090741635 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 17 |
Hb_001936_190 |
0.110616276 |
- |
- |
hypothetical protein JCGZ_17780 [Jatropha curcas] |
| 18 |
Hb_008304_010 |
0.1114035135 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 19 |
Hb_070498_010 |
0.1130025084 |
- |
- |
receptor kinase, putative [Ricinus communis] |
| 20 |
Hb_007237_020 |
0.1144145807 |
- |
- |
PREDICTED: protein trichome birefringence-like 23 [Jatropha curcas] |