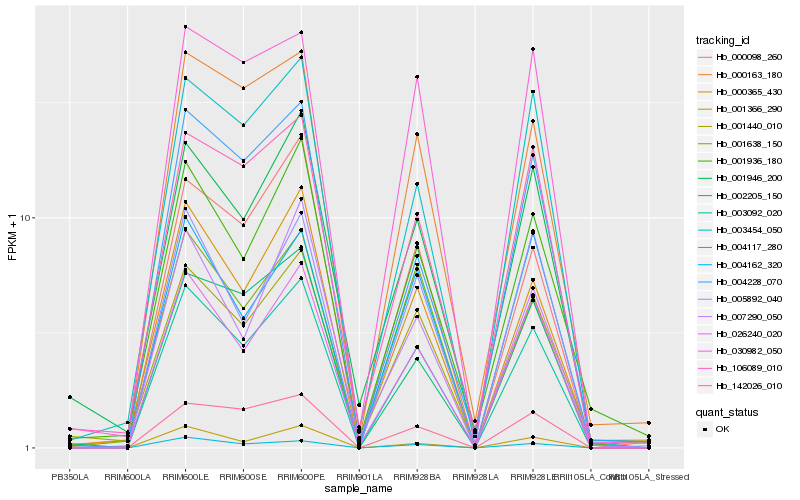
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003092_020 |
0.0 |
- |
- |
PREDICTED: heptahelical transmembrane protein 1 [Jatropha curcas] |
| 2 |
Hb_026240_020 |
0.070841196 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 3 |
Hb_000365_430 |
0.0795081374 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g08570-like [Jatropha curcas] |
| 4 |
Hb_004117_280 |
0.0867742808 |
- |
- |
hypothetical protein JCGZ_00361 [Jatropha curcas] |
| 5 |
Hb_142026_010 |
0.0937480506 |
- |
- |
PREDICTED: probable mannitol dehydrogenase [Jatropha curcas] |
| 6 |
Hb_000098_260 |
0.0949313471 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 7 |
Hb_003454_050 |
0.0972441284 |
- |
- |
PREDICTED: probable methyltransferase PMT14 [Jatropha curcas] |
| 8 |
Hb_000163_180 |
0.0972583107 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 9 |
Hb_005892_040 |
0.10010021 |
- |
- |
hypothetical protein JCGZ_02368 [Jatropha curcas] |
| 10 |
Hb_106089_010 |
0.1043543537 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 11 |
Hb_030982_050 |
0.104746631 |
- |
- |
PREDICTED: gamma-glutamyl hydrolase 2-like [Jatropha curcas] |
| 12 |
Hb_002205_150 |
0.1069770715 |
- |
- |
RING-H2 finger protein ATL3C, putative [Ricinus communis] |
| 13 |
Hb_001366_290 |
0.1075743306 |
- |
- |
Alpha-expansin 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_001638_150 |
0.1079197684 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 15 |
Hb_001936_180 |
0.109277979 |
- |
- |
PREDICTED: protein NSP-INTERACTING KINASE 3 [Jatropha curcas] |
| 16 |
Hb_001440_010 |
0.1096818494 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 1 [Populus euphratica] |
| 17 |
Hb_004228_070 |
0.1110957949 |
- |
- |
PREDICTED: uncharacterized protein LOC103338699 [Prunus mume] |
| 18 |
Hb_007290_050 |
0.1143385137 |
- |
- |
PREDICTED: uncharacterized protein LOC105629472 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001946_200 |
0.1155587673 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 20 |
Hb_004162_320 |
0.1163755945 |
- |
- |
PREDICTED: uncharacterized protein LOC105628009 isoform X1 [Jatropha curcas] |