| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
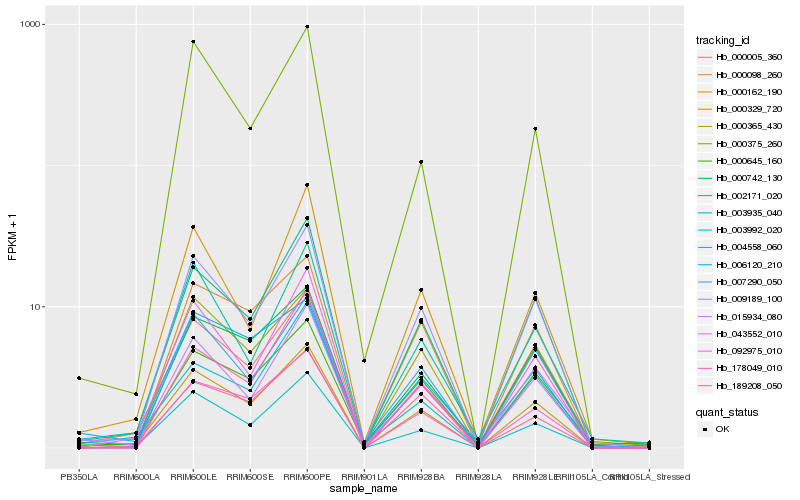
Hb_000329_720 |
0.0 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0014s05500g [Populus trichocarpa] |
| 2 |
Hb_009189_100 |
0.0639223148 |
- |
- |
PREDICTED: patellin-6 [Jatropha curcas] |
| 3 |
Hb_002171_020 |
0.0664296049 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 4 |
Hb_000005_360 |
0.0707344816 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_092975_010 |
0.0846806346 |
- |
- |
hypothetical protein JCGZ_04359 [Jatropha curcas] |
| 6 |
Hb_000365_430 |
0.0932273082 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g08570-like [Jatropha curcas] |
| 7 |
Hb_000742_130 |
0.0945794583 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g30570 [Jatropha curcas] |
| 8 |
Hb_003992_020 |
0.0948176928 |
- |
- |
PREDICTED: phosphatidylcholine:diacylglycerol cholinephosphotransferase 1-like [Jatropha curcas] |
| 9 |
Hb_015934_080 |
0.100114334 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 10 |
Hb_000098_260 |
0.1027782633 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 11 |
Hb_000645_160 |
0.1034278735 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 12 |
Hb_007290_050 |
0.1108251979 |
- |
- |
PREDICTED: uncharacterized protein LOC105629472 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000375_260 |
0.1169056509 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K4 [Manihot esculenta] |
| 14 |
Hb_189208_050 |
0.1173070802 |
- |
- |
PREDICTED: uncharacterized protein LOC105642694 isoform X2 [Jatropha curcas] |
| 15 |
Hb_003935_040 |
0.1184047498 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein SVP [Jatropha curcas] |
| 16 |
Hb_043552_010 |
0.1205729629 |
- |
- |
PREDICTED: pectinesterase/pectinesterase inhibitor PPE8B isoform X1 [Jatropha curcas] |
| 17 |
Hb_006120_210 |
0.1216375707 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 18 |
Hb_000162_190 |
0.1223353343 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 19 |
Hb_004558_060 |
0.122816316 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-4 [Jatropha curcas] |
| 20 |
Hb_178049_010 |
0.1251830783 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |