| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
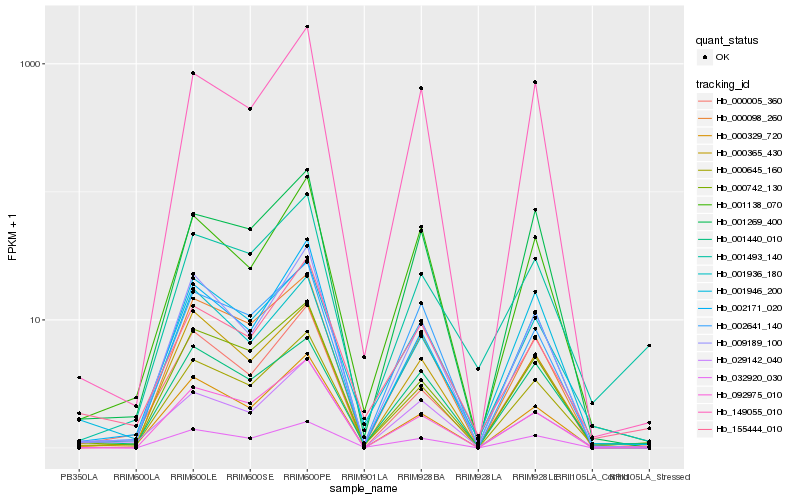
Hb_000645_160 |
0.0 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 2 |
Hb_000742_130 |
0.0790448811 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g30570 [Jatropha curcas] |
| 3 |
Hb_032920_030 |
0.081060763 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 4 |
Hb_001936_180 |
0.0900838481 |
- |
- |
PREDICTED: protein NSP-INTERACTING KINASE 3 [Jatropha curcas] |
| 5 |
Hb_000098_260 |
0.0903898937 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 6 |
Hb_001138_070 |
0.0931865663 |
- |
- |
PREDICTED: D-3-phosphoglycerate dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_149055_010 |
0.0967966979 |
- |
- |
cinnamate 4-hydroxylase [Leucaena leucocephala] |
| 8 |
Hb_001440_010 |
0.0979012484 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 1 [Populus euphratica] |
| 9 |
Hb_009189_100 |
0.0987435186 |
- |
- |
PREDICTED: patellin-6 [Jatropha curcas] |
| 10 |
Hb_001269_400 |
0.0987503324 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 11 |
Hb_002641_140 |
0.1007287629 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 12 |
Hb_001946_200 |
0.1009690591 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 13 |
Hb_000365_430 |
0.1010310376 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g08570-like [Jatropha curcas] |
| 14 |
Hb_000329_720 |
0.1034278735 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0014s05500g [Populus trichocarpa] |
| 15 |
Hb_029142_040 |
0.1095960414 |
- |
- |
replication factor C / DNA polymerase III gamma-tau subunit, putative [Ricinus communis] |
| 16 |
Hb_002171_020 |
0.1118798947 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 17 |
Hb_155444_010 |
0.1127583054 |
- |
- |
- |
| 18 |
Hb_092975_010 |
0.1152427567 |
- |
- |
hypothetical protein JCGZ_04359 [Jatropha curcas] |
| 19 |
Hb_000005_360 |
0.1170801841 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001493_140 |
0.1180297945 |
- |
- |
hypothetical protein PHAVU_010G123100g [Phaseolus vulgaris] |