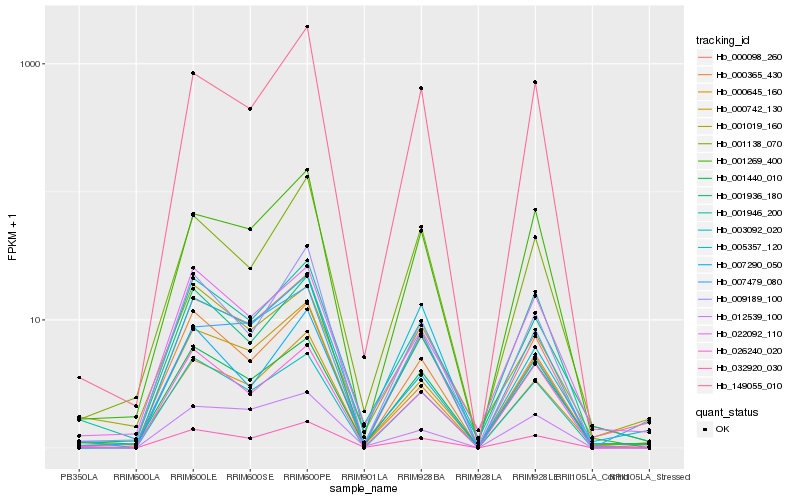
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032920_030 |
0.0 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 2 |
Hb_000645_160 |
0.081060763 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 3 |
Hb_001946_200 |
0.0947219047 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 4 |
Hb_001440_010 |
0.1009263165 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 1 [Populus euphratica] |
| 5 |
Hb_000742_130 |
0.1062691157 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g30570 [Jatropha curcas] |
| 6 |
Hb_000098_260 |
0.1135181768 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 7 |
Hb_001936_180 |
0.1148372403 |
- |
- |
PREDICTED: protein NSP-INTERACTING KINASE 3 [Jatropha curcas] |
| 8 |
Hb_022092_110 |
0.1193253183 |
- |
- |
PREDICTED: probable protein phosphatase 2C 52 [Jatropha curcas] |
| 9 |
Hb_000365_430 |
0.1222292233 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g08570-like [Jatropha curcas] |
| 10 |
Hb_149055_010 |
0.1226646329 |
- |
- |
cinnamate 4-hydroxylase [Leucaena leucocephala] |
| 11 |
Hb_003092_020 |
0.1253371625 |
- |
- |
PREDICTED: heptahelical transmembrane protein 1 [Jatropha curcas] |
| 12 |
Hb_007290_050 |
0.1258285712 |
- |
- |
PREDICTED: uncharacterized protein LOC105629472 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007479_080 |
0.1266413224 |
- |
- |
PREDICTED: uncharacterized protein LOC105649774 isoform X1 [Jatropha curcas] |
| 14 |
Hb_009189_100 |
0.1273031339 |
- |
- |
PREDICTED: patellin-6 [Jatropha curcas] |
| 15 |
Hb_001269_400 |
0.1276605825 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 16 |
Hb_026240_020 |
0.1284952769 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 17 |
Hb_001019_160 |
0.1322505729 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001138_070 |
0.1327235275 |
- |
- |
PREDICTED: D-3-phosphoglycerate dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_012539_100 |
0.133753338 |
- |
- |
PREDICTED: putative proline-rich receptor-like protein kinase PERK11 [Jatropha curcas] |
| 20 |
Hb_005357_120 |
0.1361516184 |
transcription factor |
TF Family: MYB-related |
- |