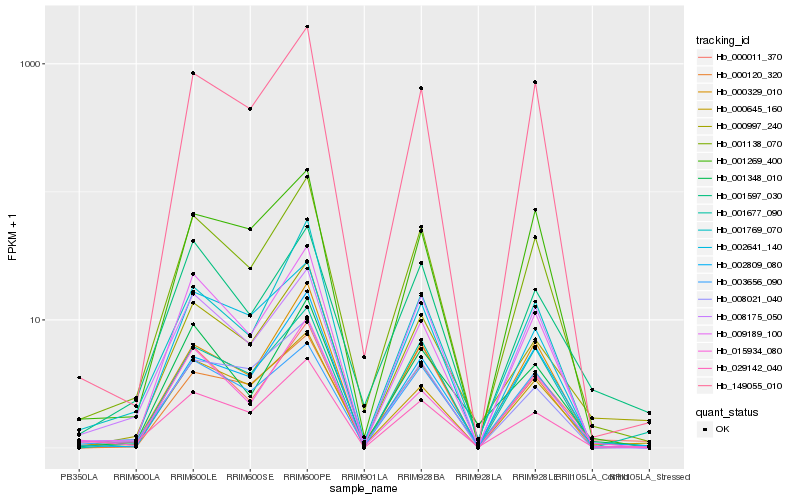
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001138_070 |
0.0 |
- |
- |
PREDICTED: D-3-phosphoglycerate dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_149055_010 |
0.0764219846 |
- |
- |
cinnamate 4-hydroxylase [Leucaena leucocephala] |
| 3 |
Hb_008175_050 |
0.0828731319 |
- |
- |
PREDICTED: formin-like protein 2 [Jatropha curcas] |
| 4 |
Hb_000645_160 |
0.0931865663 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 5 |
Hb_003656_090 |
0.093308835 |
- |
- |
PREDICTED: protein Brevis radix-like 4 [Jatropha curcas] |
| 6 |
Hb_000329_010 |
0.0942415106 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 7 |
Hb_002641_140 |
0.0963157477 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 8 |
Hb_029142_040 |
0.0967288747 |
- |
- |
replication factor C / DNA polymerase III gamma-tau subunit, putative [Ricinus communis] |
| 9 |
Hb_009189_100 |
0.0969461433 |
- |
- |
PREDICTED: patellin-6 [Jatropha curcas] |
| 10 |
Hb_000120_320 |
0.1026571532 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_001677_090 |
0.1084463071 |
- |
- |
PREDICTED: CASP-like protein 5B2 [Jatropha curcas] |
| 12 |
Hb_001269_400 |
0.1112968868 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 13 |
Hb_000011_370 |
0.1132886789 |
- |
- |
PREDICTED: uncharacterized protein LOC105631333 [Jatropha curcas] |
| 14 |
Hb_001769_070 |
0.1134939847 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_008021_040 |
0.115030117 |
- |
- |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 16 |
Hb_002809_080 |
0.1162352085 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001597_030 |
0.1166918574 |
- |
- |
PREDICTED: uncharacterized protein LOC104445128 isoform X1 [Eucalyptus grandis] |
| 18 |
Hb_000997_240 |
0.1178894286 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA4, putative [Ricinus communis] |
| 19 |
Hb_001348_010 |
0.1178978726 |
- |
- |
PREDICTED: lysM domain-containing GPI-anchored protein 1 [Jatropha curcas] |
| 20 |
Hb_015934_080 |
0.1193312757 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |