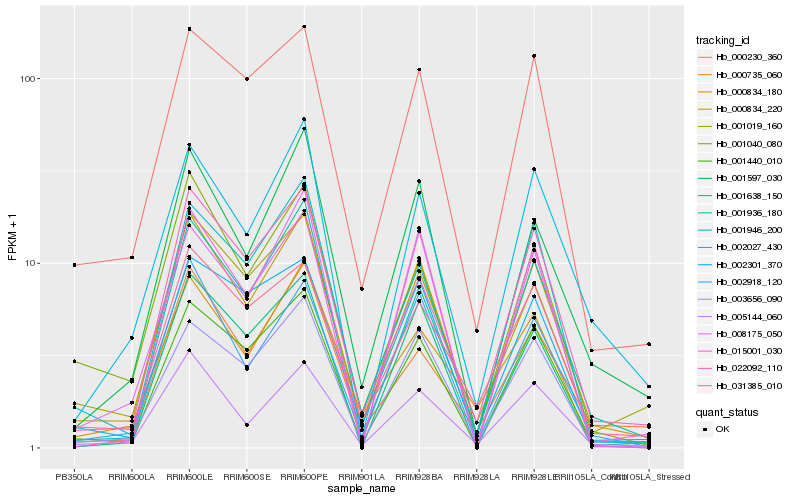
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000834_220 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 2 |
Hb_001440_010 |
0.0748607923 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 1 [Populus euphratica] |
| 3 |
Hb_005144_060 |
0.0803055254 |
- |
- |
Disease resistance family protein / LRR family protein, putative [Theobroma cacao] |
| 4 |
Hb_002918_120 |
0.0875377607 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_003656_090 |
0.0922765673 |
- |
- |
PREDICTED: protein Brevis radix-like 4 [Jatropha curcas] |
| 6 |
Hb_001936_180 |
0.0924838437 |
- |
- |
PREDICTED: protein NSP-INTERACTING KINASE 3 [Jatropha curcas] |
| 7 |
Hb_001597_030 |
0.0926125169 |
- |
- |
PREDICTED: uncharacterized protein LOC104445128 isoform X1 [Eucalyptus grandis] |
| 8 |
Hb_001019_160 |
0.0968979665 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000230_360 |
0.0981579387 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 10 |
Hb_022092_110 |
0.1043943701 |
- |
- |
PREDICTED: probable protein phosphatase 2C 52 [Jatropha curcas] |
| 11 |
Hb_031385_010 |
0.1048454332 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 12 |
Hb_001946_200 |
0.1055366951 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 13 |
Hb_001040_080 |
0.1066389105 |
- |
- |
sphingolipid delta 4 desaturase/C-4 hydroxylase protein des2, putative [Ricinus communis] |
| 14 |
Hb_015001_030 |
0.1076354047 |
- |
- |
inosine-uridine preferring nucleoside hydrolase family protein [Populus trichocarpa] |
| 15 |
Hb_002301_370 |
0.110856098 |
- |
- |
hypothetical protein JCGZ_21454 [Jatropha curcas] |
| 16 |
Hb_002027_430 |
0.1126866921 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 17 |
Hb_001638_150 |
0.1128632743 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 18 |
Hb_000735_060 |
0.1168533048 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 19 |
Hb_008175_050 |
0.117160053 |
- |
- |
PREDICTED: formin-like protein 2 [Jatropha curcas] |
| 20 |
Hb_000834_180 |
0.1174147748 |
- |
- |
PREDICTED: kinesin-13A-like isoform X1 [Jatropha curcas] |