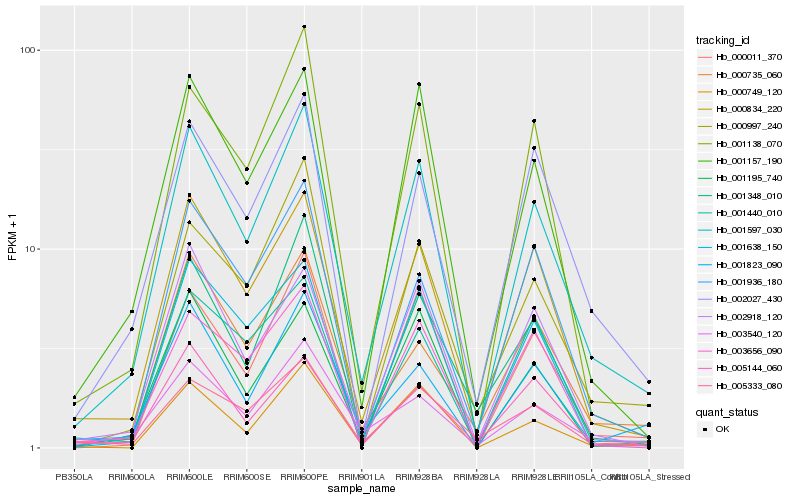
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001597_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104445128 isoform X1 [Eucalyptus grandis] |
| 2 |
Hb_000834_220 |
0.0926125169 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 3 |
Hb_001157_190 |
0.1055979985 |
- |
- |
PREDICTED: protein YLS3 [Jatropha curcas] |
| 4 |
Hb_000011_370 |
0.1128389718 |
- |
- |
PREDICTED: uncharacterized protein LOC105631333 [Jatropha curcas] |
| 5 |
Hb_000749_120 |
0.1145167051 |
- |
- |
PREDICTED: uncharacterized protein LOC105644255 isoform X2 [Jatropha curcas] |
| 6 |
Hb_003540_120 |
0.1146906606 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 7 |
Hb_001138_070 |
0.1166918574 |
- |
- |
PREDICTED: D-3-phosphoglycerate dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_001823_090 |
0.1167634334 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000997_240 |
0.119368518 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA4, putative [Ricinus communis] |
| 10 |
Hb_002027_430 |
0.1198655476 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 11 |
Hb_002918_120 |
0.1217619642 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_001348_010 |
0.1235802554 |
- |
- |
PREDICTED: lysM domain-containing GPI-anchored protein 1 [Jatropha curcas] |
| 13 |
Hb_001195_740 |
0.1237580685 |
transcription factor |
TF Family: LIM |
PREDICTED: protein DA1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001936_180 |
0.1254786696 |
- |
- |
PREDICTED: protein NSP-INTERACTING KINASE 3 [Jatropha curcas] |
| 15 |
Hb_005333_080 |
0.1290955904 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003656_090 |
0.1317549211 |
- |
- |
PREDICTED: protein Brevis radix-like 4 [Jatropha curcas] |
| 17 |
Hb_000735_060 |
0.132461451 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 18 |
Hb_001638_150 |
0.1328956077 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 19 |
Hb_001440_010 |
0.1329788827 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 1 [Populus euphratica] |
| 20 |
Hb_005144_060 |
0.1340391206 |
- |
- |
Disease resistance family protein / LRR family protein, putative [Theobroma cacao] |