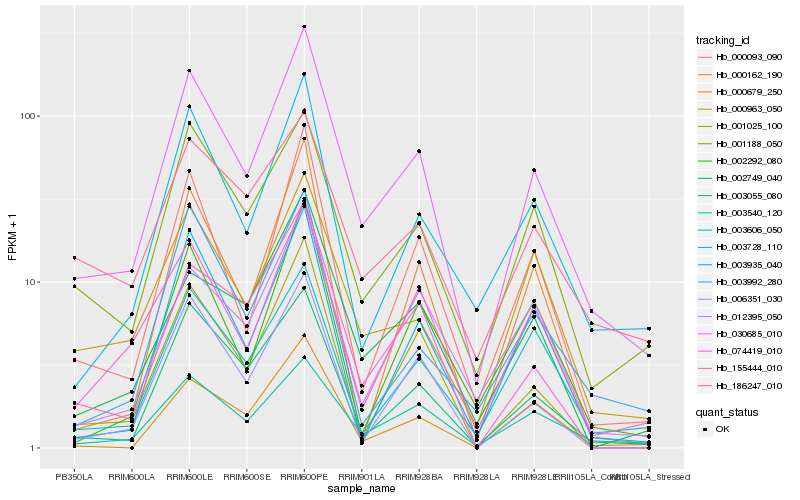
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030685_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_003540_120 |
0.1125552126 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 3 |
Hb_000093_090 |
0.1227281691 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 3 [Populus euphratica] |
| 4 |
Hb_003728_110 |
0.123061671 |
- |
- |
PREDICTED: calmodulin-like protein 3 [Populus euphratica] |
| 5 |
Hb_001188_050 |
0.1239924757 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003055_080 |
0.1280956425 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002749_040 |
0.1369552698 |
- |
- |
Ribosomal protein L6 family protein [Theobroma cacao] |
| 8 |
Hb_000963_050 |
0.1373302503 |
- |
- |
PREDICTED: vegetative cell wall protein gp1-like [Jatropha curcas] |
| 9 |
Hb_155444_010 |
0.1380068588 |
- |
- |
- |
| 10 |
Hb_074419_010 |
0.1400122904 |
- |
- |
PREDICTED: uncharacterized protein LOC105637596 [Jatropha curcas] |
| 11 |
Hb_001025_100 |
0.140718905 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 12 |
Hb_012395_050 |
0.1439063125 |
- |
- |
PREDICTED: uncharacterized protein LOC105632180 [Jatropha curcas] |
| 13 |
Hb_003606_050 |
0.1466599689 |
- |
- |
PREDICTED: beta-galactosidase 10 [Jatropha curcas] |
| 14 |
Hb_000679_250 |
0.1485947606 |
- |
- |
PREDICTED: uncharacterized protein LOC105637632 isoform X1 [Jatropha curcas] |
| 15 |
Hb_006351_030 |
0.1508225225 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MARCH8-like [Jatropha curcas] |
| 16 |
Hb_003935_040 |
0.152935896 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein SVP [Jatropha curcas] |
| 17 |
Hb_000162_190 |
0.1531313921 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 18 |
Hb_003992_280 |
0.1542740869 |
- |
- |
PREDICTED: uncharacterized protein LOC105117987 [Populus euphratica] |
| 19 |
Hb_186247_010 |
0.1554645533 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase-like [Pyrus x bretschneideri] |
| 20 |
Hb_002292_080 |
0.1555169527 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |