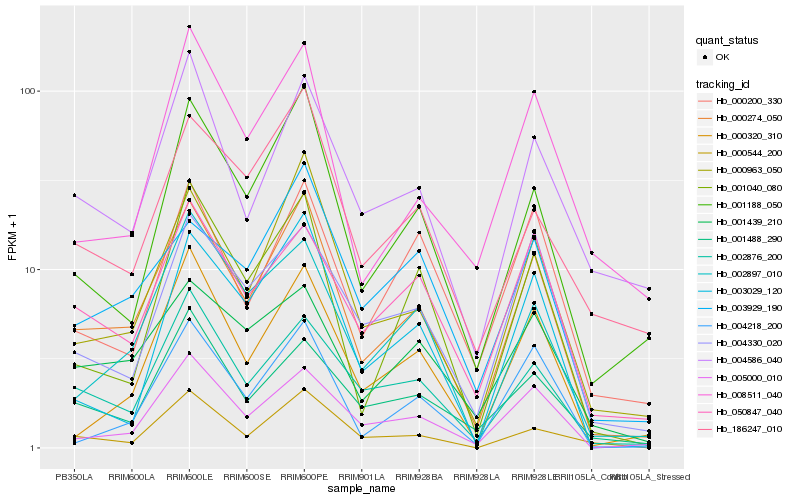
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000274_050 |
0.0 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 2 |
Hb_000963_050 |
0.1015329452 |
- |
- |
PREDICTED: vegetative cell wall protein gp1-like [Jatropha curcas] |
| 3 |
Hb_001188_050 |
0.1258821212 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005000_010 |
0.1282494517 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002876_200 |
0.1303151848 |
- |
- |
PREDICTED: uncharacterized protein LOC105641209 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001040_080 |
0.1428791742 |
- |
- |
sphingolipid delta 4 desaturase/C-4 hydroxylase protein des2, putative [Ricinus communis] |
| 7 |
Hb_001439_210 |
0.1436310272 |
- |
- |
EXS family protein [Populus trichocarpa] |
| 8 |
Hb_050847_040 |
0.1442130811 |
- |
- |
PREDICTED: uncharacterized protein LOC105649141 [Jatropha curcas] |
| 9 |
Hb_004586_040 |
0.1446337325 |
transcription factor |
TF Family: MIKC |
K-box region and MADS-box transcription factor family protein [Theobroma cacao] |
| 10 |
Hb_002897_010 |
0.146681517 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g58300 [Jatropha curcas] |
| 11 |
Hb_000320_310 |
0.1468808216 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C [Jatropha curcas] |
| 12 |
Hb_004218_200 |
0.1476073102 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 13 |
Hb_000544_200 |
0.1477779607 |
- |
- |
PREDICTED: uncharacterized protein LOC105647171 [Jatropha curcas] |
| 14 |
Hb_003929_190 |
0.1496980699 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g18500 [Jatropha curcas] |
| 15 |
Hb_004330_020 |
0.1498717296 |
- |
- |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 16 |
Hb_001488_290 |
0.1504109584 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 17 |
Hb_003029_120 |
0.1511253311 |
- |
- |
PREDICTED: uncharacterized protein LOC105642638 [Jatropha curcas] |
| 18 |
Hb_186247_010 |
0.1518463312 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase-like [Pyrus x bretschneideri] |
| 19 |
Hb_008511_040 |
0.1542355466 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 20 |
Hb_000200_330 |
0.1592950374 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |