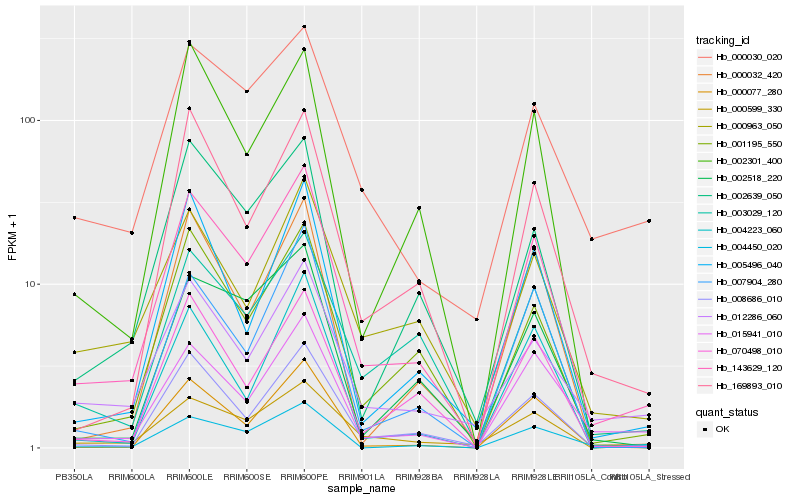
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_143629_120 |
0.0 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 2 |
Hb_008686_010 |
0.1002758764 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001195_550 |
0.1128580417 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000599_330 |
0.1133964532 |
- |
- |
hypothetical protein JCGZ_17661 [Jatropha curcas] |
| 5 |
Hb_015941_010 |
0.118987932 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 6 |
Hb_169893_010 |
0.1268853455 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 7 |
Hb_012286_060 |
0.1286193438 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_000963_050 |
0.1326596035 |
- |
- |
PREDICTED: vegetative cell wall protein gp1-like [Jatropha curcas] |
| 9 |
Hb_002639_050 |
0.13349406 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 10 |
Hb_004450_020 |
0.1345909552 |
- |
- |
serine-threonine protein kinase, putative [Ricinus communis] |
| 11 |
Hb_005496_040 |
0.135133797 |
- |
- |
PREDICTED: uncharacterized protein LOC105630864 [Jatropha curcas] |
| 12 |
Hb_070498_010 |
0.1352127997 |
- |
- |
receptor kinase, putative [Ricinus communis] |
| 13 |
Hb_007904_280 |
0.135214607 |
- |
- |
PREDICTED: uncharacterized protein LOC105644987 [Jatropha curcas] |
| 14 |
Hb_002301_400 |
0.1383578608 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 15 |
Hb_000077_280 |
0.1384799539 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_003029_120 |
0.1393921767 |
- |
- |
PREDICTED: uncharacterized protein LOC105642638 [Jatropha curcas] |
| 17 |
Hb_000030_020 |
0.1394438451 |
- |
- |
PREDICTED: uncharacterized protein LOC105123549 [Populus euphratica] |
| 18 |
Hb_002518_220 |
0.1402784628 |
- |
- |
PREDICTED: uncharacterized protein LOC105631001 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000032_420 |
0.1432620233 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 20 |
Hb_004223_060 |
0.1433040051 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |