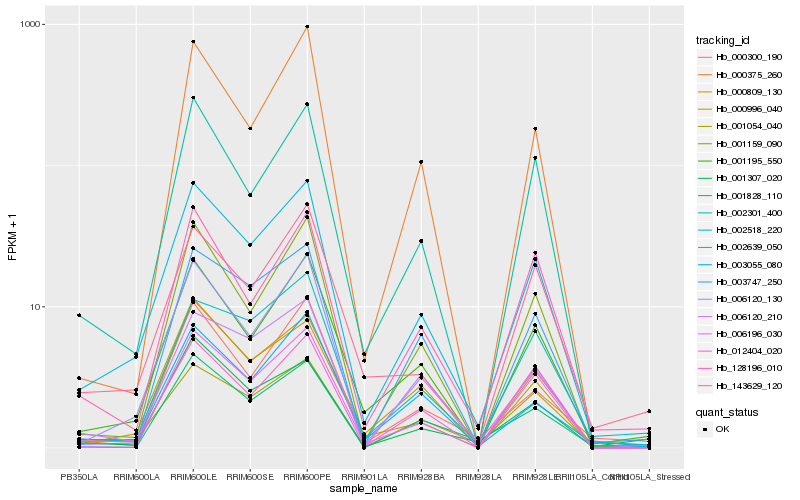
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002639_050 |
0.0 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 2 |
Hb_001195_550 |
0.0964281954 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001054_040 |
0.102125499 |
- |
- |
hypothetical protein JCGZ_25310 [Jatropha curcas] |
| 4 |
Hb_002301_400 |
0.1117226469 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 5 |
Hb_001159_090 |
0.1133597263 |
- |
- |
PREDICTED: methylsterol monooxygenase 1-1-like [Jatropha curcas] |
| 6 |
Hb_006196_030 |
0.1149667262 |
- |
- |
PREDICTED: uncharacterized protein LOC105650679 [Jatropha curcas] |
| 7 |
Hb_000300_190 |
0.1157230594 |
- |
- |
PREDICTED: uncharacterized protein LOC105632566 [Jatropha curcas] |
| 8 |
Hb_000996_040 |
0.1160136575 |
- |
- |
hypothetical protein POPTR_0003s12500g [Populus trichocarpa] |
| 9 |
Hb_006120_210 |
0.1169678054 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 10 |
Hb_006120_130 |
0.1173741827 |
- |
- |
- |
| 11 |
Hb_003055_080 |
0.1222528956 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003747_250 |
0.1241353696 |
- |
- |
PREDICTED: potassium transporter 7 isoform X2 [Elaeis guineensis] |
| 13 |
Hb_000809_130 |
0.124205057 |
- |
- |
PREDICTED: peroxidase 10 isoform X3 [Jatropha curcas] |
| 14 |
Hb_001828_110 |
0.1261543766 |
- |
- |
PREDICTED: lysine histidine transporter 1-like [Tarenaya hassleriana] |
| 15 |
Hb_002518_220 |
0.1266050452 |
- |
- |
PREDICTED: uncharacterized protein LOC105631001 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000375_260 |
0.1304050684 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K4 [Manihot esculenta] |
| 17 |
Hb_001307_020 |
0.1314113715 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 18 |
Hb_012404_020 |
0.1314559143 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_143629_120 |
0.13349406 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 20 |
Hb_128196_010 |
0.1350366841 |
- |
- |
conserved hypothetical protein [Ricinus communis] |