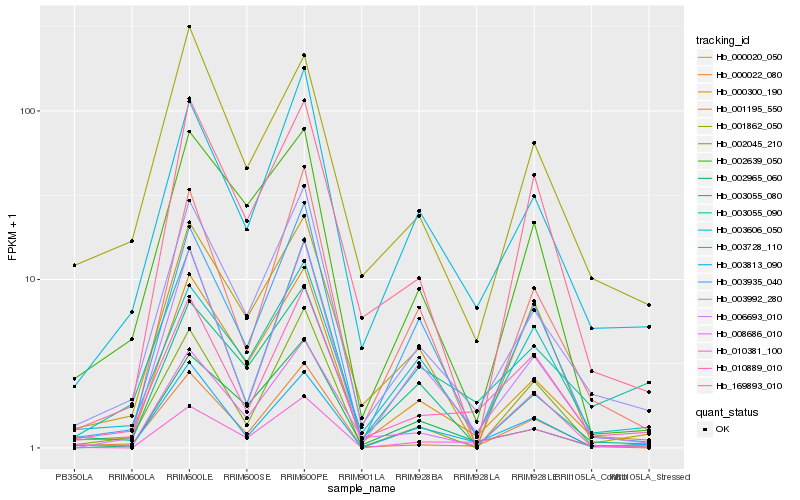
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003992_280 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105117987 [Populus euphratica] |
| 2 |
Hb_000300_190 |
0.0626902012 |
- |
- |
PREDICTED: uncharacterized protein LOC105632566 [Jatropha curcas] |
| 3 |
Hb_003728_110 |
0.0836551457 |
- |
- |
PREDICTED: calmodulin-like protein 3 [Populus euphratica] |
| 4 |
Hb_003055_080 |
0.1199931873 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002965_060 |
0.1234904137 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 6 |
Hb_001195_550 |
0.1279829587 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000020_050 |
0.1302015047 |
- |
- |
PREDICTED: expansin-A6 [Jatropha curcas] |
| 8 |
Hb_003935_040 |
0.1303819992 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein SVP [Jatropha curcas] |
| 9 |
Hb_000022_080 |
0.1322577439 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 10 |
Hb_010889_010 |
0.133492133 |
- |
- |
PREDICTED: myosin-12 [Jatropha curcas] |
| 11 |
Hb_006693_010 |
0.1335180538 |
- |
- |
PREDICTED: uncharacterized protein LOC105628875 [Jatropha curcas] |
| 12 |
Hb_008686_010 |
0.1345967159 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003813_090 |
0.1380693393 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 14 |
Hb_001862_050 |
0.1385997926 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 18-like [Jatropha curcas] |
| 15 |
Hb_003055_090 |
0.1392150629 |
- |
- |
PREDICTED: uncharacterized protein LOC105649490 [Jatropha curcas] |
| 16 |
Hb_002639_050 |
0.1419676592 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 17 |
Hb_002045_210 |
0.1435176192 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 18 |
Hb_010381_100 |
0.1442037844 |
- |
- |
Phospholipase D epsilon [Morus notabilis] |
| 19 |
Hb_169893_010 |
0.1446051344 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 20 |
Hb_003606_050 |
0.1457429666 |
- |
- |
PREDICTED: beta-galactosidase 10 [Jatropha curcas] |