| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
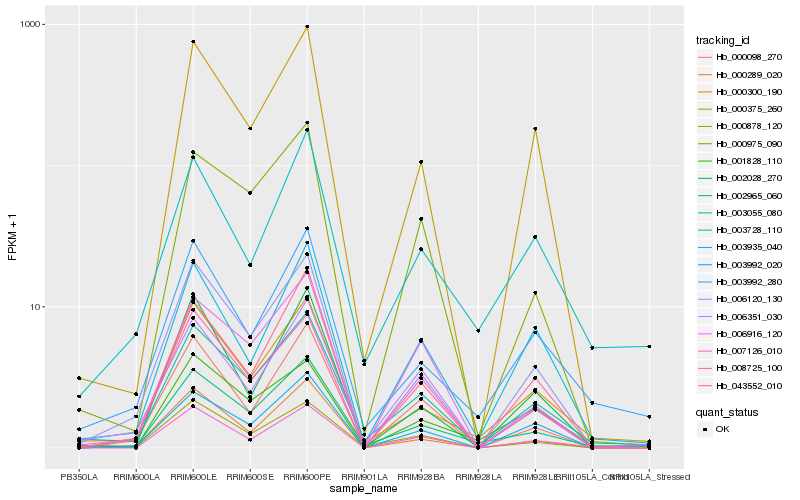
Hb_002965_060 |
0.0 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 2 |
Hb_000300_190 |
0.0975365118 |
- |
- |
PREDICTED: uncharacterized protein LOC105632566 [Jatropha curcas] |
| 3 |
Hb_003055_080 |
0.1011431784 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003992_280 |
0.1234904137 |
- |
- |
PREDICTED: uncharacterized protein LOC105117987 [Populus euphratica] |
| 5 |
Hb_006120_130 |
0.1333045755 |
- |
- |
- |
| 6 |
Hb_000375_260 |
0.1376113877 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K4 [Manihot esculenta] |
| 7 |
Hb_003728_110 |
0.140255455 |
- |
- |
PREDICTED: calmodulin-like protein 3 [Populus euphratica] |
| 8 |
Hb_000289_020 |
0.1423645085 |
- |
- |
hypothetical protein CICLE_v10009323mg [Citrus clementina] |
| 9 |
Hb_000975_090 |
0.1423868766 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 10 |
Hb_003992_020 |
0.1434508433 |
- |
- |
PREDICTED: phosphatidylcholine:diacylglycerol cholinephosphotransferase 1-like [Jatropha curcas] |
| 11 |
Hb_043552_010 |
0.1445670504 |
- |
- |
PREDICTED: pectinesterase/pectinesterase inhibitor PPE8B isoform X1 [Jatropha curcas] |
| 12 |
Hb_001828_110 |
0.1460692994 |
- |
- |
PREDICTED: lysine histidine transporter 1-like [Tarenaya hassleriana] |
| 13 |
Hb_000098_270 |
0.1476755249 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein [Jatropha curcas] |
| 14 |
Hb_008725_100 |
0.1479039402 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 15 |
Hb_007126_010 |
0.1488189905 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 16 |
Hb_000878_120 |
0.1500537331 |
- |
- |
PREDICTED: protein downstream neighbor of Son [Jatropha curcas] |
| 17 |
Hb_006916_120 |
0.1522119984 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 1 [Jatropha curcas] |
| 18 |
Hb_003935_040 |
0.154735161 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein SVP [Jatropha curcas] |
| 19 |
Hb_002028_270 |
0.1551895025 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 20 |
Hb_006351_030 |
0.1564771906 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MARCH8-like [Jatropha curcas] |