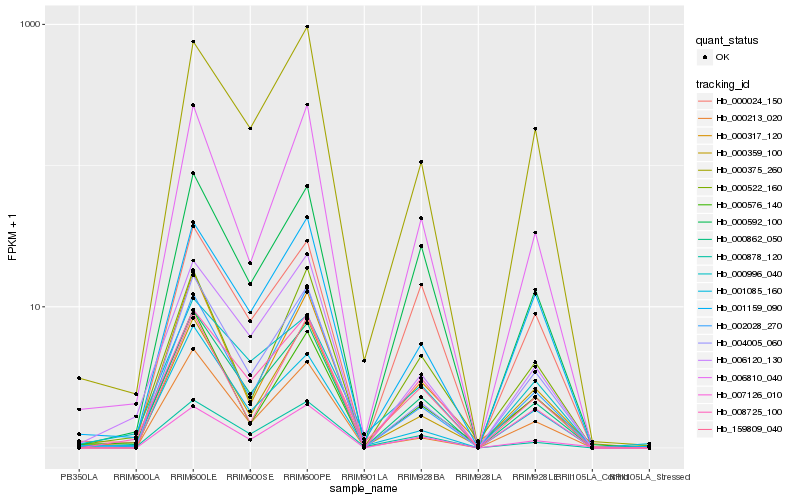
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000862_050 |
0.0 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 2 |
Hb_008725_100 |
0.077251909 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 3 |
Hb_000592_100 |
0.085405206 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 4 |
Hb_004005_060 |
0.0913532918 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 5 |
Hb_006120_130 |
0.0992095283 |
- |
- |
- |
| 6 |
Hb_006810_040 |
0.1009510711 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 7 |
Hb_007126_010 |
0.103198239 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 8 |
Hb_001159_090 |
0.1104890821 |
- |
- |
PREDICTED: methylsterol monooxygenase 1-1-like [Jatropha curcas] |
| 9 |
Hb_000375_260 |
0.1108384924 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K4 [Manihot esculenta] |
| 10 |
Hb_000996_040 |
0.1117578825 |
- |
- |
hypothetical protein POPTR_0003s12500g [Populus trichocarpa] |
| 11 |
Hb_000576_140 |
0.1118753618 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 12 |
Hb_000878_120 |
0.1120374025 |
- |
- |
PREDICTED: protein downstream neighbor of Son [Jatropha curcas] |
| 13 |
Hb_000317_120 |
0.1129252065 |
- |
- |
PREDICTED: uncharacterized protein LOC105647000 [Jatropha curcas] |
| 14 |
Hb_002028_270 |
0.1132142333 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 15 |
Hb_000213_020 |
0.1200662615 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001085_160 |
0.1221284941 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 17 |
Hb_159809_040 |
0.1222744676 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000522_160 |
0.1223487007 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 19 |
Hb_000359_100 |
0.122872374 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000024_150 |
0.1230289123 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |