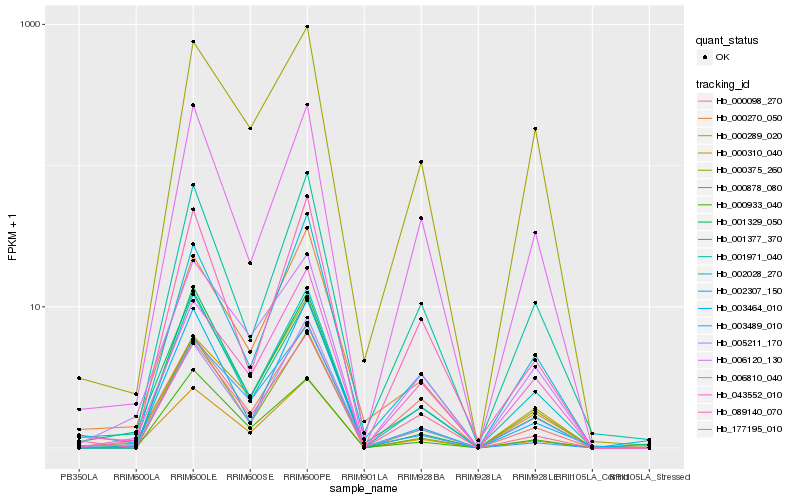
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000289_020 |
0.0 |
- |
- |
hypothetical protein CICLE_v10009323mg [Citrus clementina] |
| 2 |
Hb_006120_130 |
0.0860510006 |
- |
- |
- |
| 3 |
Hb_002028_270 |
0.0894422754 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 4 |
Hb_000098_270 |
0.0931140202 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein [Jatropha curcas] |
| 5 |
Hb_005211_170 |
0.0996830395 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 6 |
Hb_043552_010 |
0.1079804429 |
- |
- |
PREDICTED: pectinesterase/pectinesterase inhibitor PPE8B isoform X1 [Jatropha curcas] |
| 7 |
Hb_003464_010 |
0.1117460165 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000270_050 |
0.1118652979 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FEI 2 [Jatropha curcas] |
| 9 |
Hb_001377_370 |
0.1122455116 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 10 |
Hb_177195_010 |
0.1135730927 |
- |
- |
Leucine-rich repeat protein kinase family protein isoform 3, partial [Theobroma cacao] |
| 11 |
Hb_000933_040 |
0.1158133406 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 12 |
Hb_002307_150 |
0.1172032058 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA4-like [Jatropha curcas] |
| 13 |
Hb_003489_010 |
0.117433023 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Populus euphratica] |
| 14 |
Hb_000310_040 |
0.1194320937 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 3 [Jatropha curcas] |
| 15 |
Hb_001971_040 |
0.1200988082 |
- |
- |
PREDICTED: beta-glucosidase 44-like [Jatropha curcas] |
| 16 |
Hb_006810_040 |
0.1212670995 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 17 |
Hb_089140_070 |
0.1242251297 |
- |
- |
PREDICTED: uncharacterized protein LOC105637961 [Jatropha curcas] |
| 18 |
Hb_001329_050 |
0.1244885426 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000878_080 |
0.1268618785 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 20 |
Hb_000375_260 |
0.1298359982 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K4 [Manihot esculenta] |