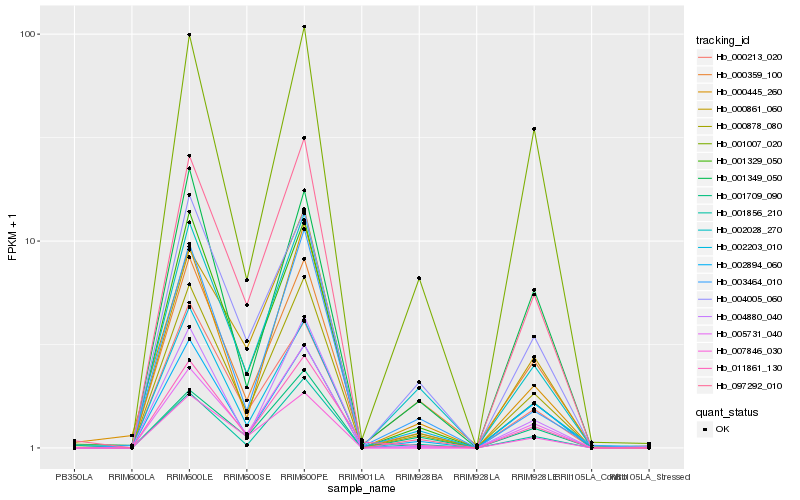
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000878_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 2 |
Hb_002028_270 |
0.0753303082 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 3 |
Hb_097292_010 |
0.0835084647 |
- |
- |
PREDICTED: uncharacterized protein LOC105643999 [Jatropha curcas] |
| 4 |
Hb_001856_210 |
0.0853203533 |
- |
- |
PREDICTED: uncharacterized protein LOC105644171 [Jatropha curcas] |
| 5 |
Hb_002894_060 |
0.0907612193 |
- |
- |
respiratory burst oxidase D [Manihot esculenta] |
| 6 |
Hb_002203_010 |
0.0913310082 |
- |
- |
PREDICTED: kinesin-like protein KIF18B isoform X1 [Vitis vinifera] |
| 7 |
Hb_004005_060 |
0.0929947475 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 8 |
Hb_004880_040 |
0.0954158391 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X1 [Jatropha curcas] |
| 9 |
Hb_011861_130 |
0.0967426585 |
- |
- |
PREDICTED: uncharacterized protein LOC105634795 [Jatropha curcas] |
| 10 |
Hb_000359_100 |
0.0982064549 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000213_020 |
0.098322745 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001329_050 |
0.0999644098 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007846_030 |
0.1020711869 |
- |
- |
Uncharacterized protein TCM_021839 [Theobroma cacao] |
| 14 |
Hb_005731_040 |
0.1036621316 |
- |
- |
PREDICTED: glutaredoxin-C1-like [Jatropha curcas] |
| 15 |
Hb_000445_260 |
0.104246888 |
- |
- |
PREDICTED: uncharacterized protein LOC105127729 [Populus euphratica] |
| 16 |
Hb_001349_050 |
0.1058433342 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 17 |
Hb_003464_010 |
0.106485824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001007_020 |
0.1102395269 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000861_060 |
0.1112395838 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 20 |
Hb_001709_090 |
0.1112462772 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |