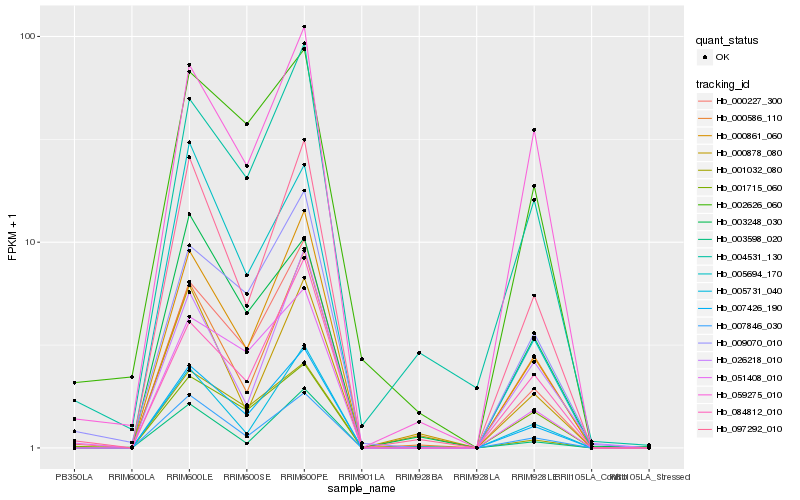
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007426_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103325200 [Prunus mume] |
| 2 |
Hb_000227_300 |
0.065862847 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007846_030 |
0.0685023 |
- |
- |
Uncharacterized protein TCM_021839 [Theobroma cacao] |
| 4 |
Hb_000861_060 |
0.0744745424 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 5 |
Hb_097292_010 |
0.0776891234 |
- |
- |
PREDICTED: uncharacterized protein LOC105643999 [Jatropha curcas] |
| 6 |
Hb_005731_040 |
0.1043019165 |
- |
- |
PREDICTED: glutaredoxin-C1-like [Jatropha curcas] |
| 7 |
Hb_051408_010 |
0.107826356 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 8 |
Hb_000586_110 |
0.1100589757 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 9 |
Hb_009070_010 |
0.1164386581 |
- |
- |
Spermidine synthase, putative [Ricinus communis] |
| 10 |
Hb_005694_170 |
0.1192914683 |
- |
- |
PREDICTED: uncharacterized protein At3g49140 isoform X2 [Jatropha curcas] |
| 11 |
Hb_084812_010 |
0.1200608741 |
- |
- |
PREDICTED: putative RING-H2 finger protein ATL21B [Vitis vinifera] |
| 12 |
Hb_001715_060 |
0.122385477 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 1 [Jatropha curcas] |
| 13 |
Hb_001032_080 |
0.1234229708 |
- |
- |
Ankyrin repeat protein [Medicago truncatula] |
| 14 |
Hb_059275_010 |
0.1236895227 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 15 |
Hb_004531_130 |
0.1306130846 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_003248_030 |
0.13392247 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_026218_010 |
0.1365661576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003598_020 |
0.1380541783 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12 [Jatropha curcas] |
| 19 |
Hb_000878_080 |
0.1400328262 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 20 |
Hb_002626_060 |
0.1448075612 |
- |
- |
hypothetical protein JCGZ_27059 [Jatropha curcas] |