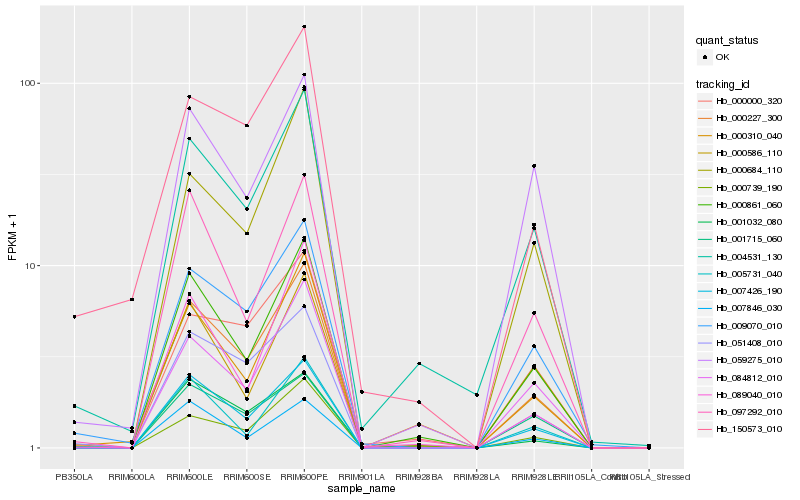
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000227_300 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007426_190 |
0.065862847 |
- |
- |
PREDICTED: uncharacterized protein LOC103325200 [Prunus mume] |
| 3 |
Hb_000861_060 |
0.076641185 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 4 |
Hb_051408_010 |
0.0853780516 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 5 |
Hb_009070_010 |
0.086536473 |
- |
- |
Spermidine synthase, putative [Ricinus communis] |
| 6 |
Hb_004531_130 |
0.1067020625 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_097292_010 |
0.1071219322 |
- |
- |
PREDICTED: uncharacterized protein LOC105643999 [Jatropha curcas] |
| 8 |
Hb_084812_010 |
0.1082252908 |
- |
- |
PREDICTED: putative RING-H2 finger protein ATL21B [Vitis vinifera] |
| 9 |
Hb_001032_080 |
0.1149585051 |
- |
- |
Ankyrin repeat protein [Medicago truncatula] |
| 10 |
Hb_000739_190 |
0.1163902551 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000310_040 |
0.1205624067 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 3 [Jatropha curcas] |
| 12 |
Hb_000684_110 |
0.120841658 |
- |
- |
hypothetical protein POPTR_0006s13250g [Populus trichocarpa] |
| 13 |
Hb_007846_030 |
0.1217330503 |
- |
- |
Uncharacterized protein TCM_021839 [Theobroma cacao] |
| 14 |
Hb_089040_010 |
0.1296375524 |
- |
- |
Glycosyl hydrolase family protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_005731_040 |
0.1301550093 |
- |
- |
PREDICTED: glutaredoxin-C1-like [Jatropha curcas] |
| 16 |
Hb_000000_320 |
0.1326080596 |
- |
- |
PREDICTED: calcium uniporter protein 4, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000586_110 |
0.1334398829 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 18 |
Hb_059275_010 |
0.1351698697 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 19 |
Hb_150573_010 |
0.1362580778 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |
| 20 |
Hb_001715_060 |
0.1372837815 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 1 [Jatropha curcas] |