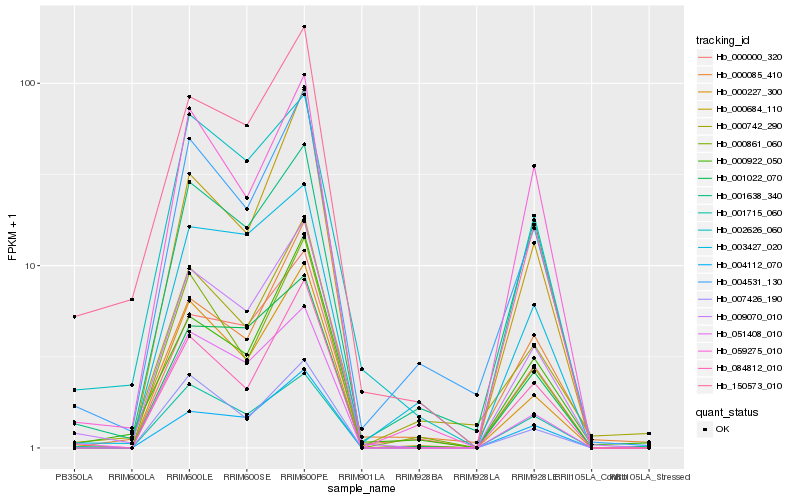
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009070_010 |
0.0 |
- |
- |
Spermidine synthase, putative [Ricinus communis] |
| 2 |
Hb_000227_300 |
0.086536473 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_051408_010 |
0.0940698078 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 4 |
Hb_004531_130 |
0.0999103726 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_000000_320 |
0.1059689023 |
- |
- |
PREDICTED: calcium uniporter protein 4, mitochondrial [Jatropha curcas] |
| 6 |
Hb_002626_060 |
0.1071754728 |
- |
- |
hypothetical protein JCGZ_27059 [Jatropha curcas] |
| 7 |
Hb_004112_070 |
0.1089962096 |
- |
- |
- |
| 8 |
Hb_150573_010 |
0.1107801921 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |
| 9 |
Hb_084812_010 |
0.1126212777 |
- |
- |
PREDICTED: putative RING-H2 finger protein ATL21B [Vitis vinifera] |
| 10 |
Hb_001715_060 |
0.1151511718 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 1 [Jatropha curcas] |
| 11 |
Hb_001022_070 |
0.1162561812 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 12 |
Hb_007426_190 |
0.1164386581 |
- |
- |
PREDICTED: uncharacterized protein LOC103325200 [Prunus mume] |
| 13 |
Hb_059275_010 |
0.122134682 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 14 |
Hb_000684_110 |
0.1249823563 |
- |
- |
hypothetical protein POPTR_0006s13250g [Populus trichocarpa] |
| 15 |
Hb_000922_050 |
0.1295427696 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 16 |
Hb_000861_060 |
0.1295443654 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 17 |
Hb_000085_410 |
0.131530005 |
- |
- |
hypothetical protein POPTR_0011s09040g [Populus trichocarpa] |
| 18 |
Hb_001638_340 |
0.1327147041 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000742_290 |
0.1332860169 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 20 |
Hb_003427_020 |
0.1337301016 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X1 [Jatropha curcas] |