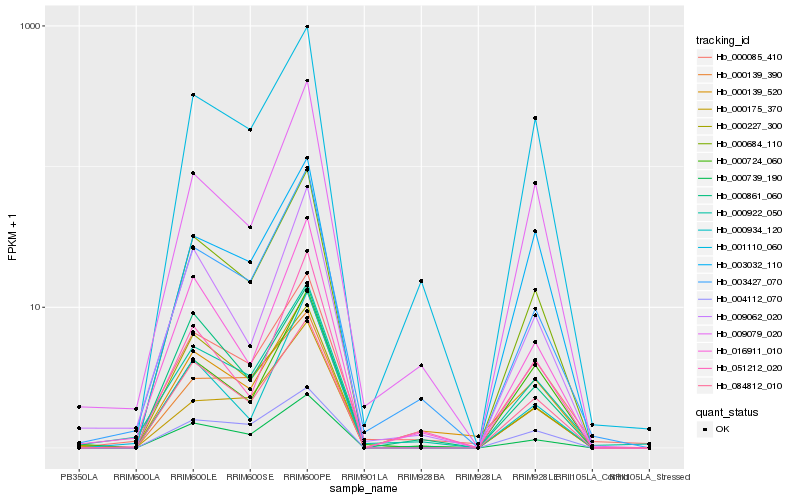
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000684_110 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s13250g [Populus trichocarpa] |
| 2 |
Hb_084812_010 |
0.0512623348 |
- |
- |
PREDICTED: putative RING-H2 finger protein ATL21B [Vitis vinifera] |
| 3 |
Hb_000739_190 |
0.0858759881 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_004112_070 |
0.091462797 |
- |
- |
- |
| 5 |
Hb_001110_060 |
0.0949538957 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 6 |
Hb_003427_070 |
0.0961687636 |
- |
- |
hypothetical protein POPTR_0067s00310g [Populus trichocarpa] |
| 7 |
Hb_000139_390 |
0.096642748 |
- |
- |
- |
| 8 |
Hb_016911_010 |
0.106516892 |
- |
- |
hypothetical protein POPTR_0010s15010g [Populus trichocarpa] |
| 9 |
Hb_000922_050 |
0.1086565049 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 10 |
Hb_000934_120 |
0.1139118851 |
- |
- |
PREDICTED: tumor necrosis factor ligand superfamily member 6 [Populus euphratica] |
| 11 |
Hb_000085_410 |
0.1169108305 |
- |
- |
hypothetical protein POPTR_0011s09040g [Populus trichocarpa] |
| 12 |
Hb_009062_020 |
0.1171864925 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP20-2, chloroplastic [Cucumis melo] |
| 13 |
Hb_000139_520 |
0.1183625612 |
- |
- |
EF-hand calcium-binding domain-containing protein 4A [Theobroma cacao] |
| 14 |
Hb_009079_020 |
0.1195125742 |
- |
- |
PREDICTED: protein WALLS ARE THIN 1 [Jatropha curcas] |
| 15 |
Hb_003032_110 |
0.1203842914 |
- |
- |
- |
| 16 |
Hb_000724_060 |
0.1207773405 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 17 |
Hb_000227_300 |
0.120841658 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000175_370 |
0.1211132279 |
- |
- |
hypothetical protein EUGRSUZ_F03862 [Eucalyptus grandis] |
| 19 |
Hb_051212_020 |
0.1220934394 |
- |
- |
PREDICTED: 24-methylenesterol C-methyltransferase 2-like [Jatropha curcas] |
| 20 |
Hb_000861_060 |
0.1240215577 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |