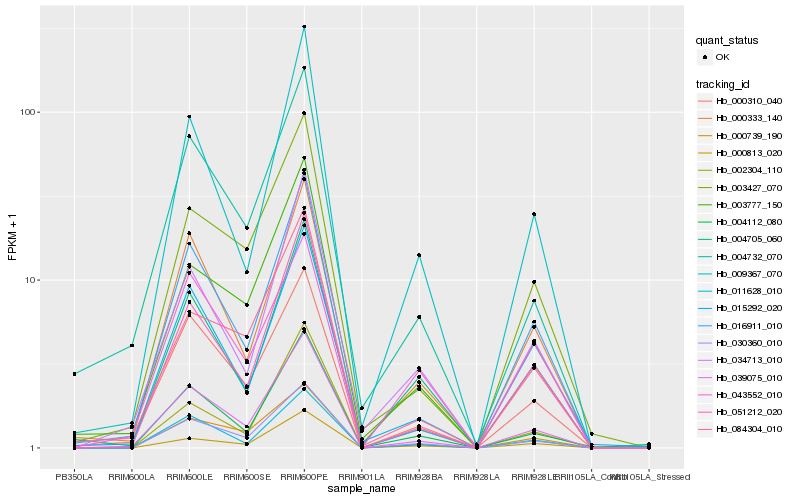
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030360_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_039075_010 |
0.0544976084 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 3 |
Hb_004112_080 |
0.0782716868 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like tyrosine-protein kinase At2g41820 [Jatropha curcas] |
| 4 |
Hb_000739_190 |
0.0805040942 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 5 |
Hb_000310_040 |
0.0873562964 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 3 [Jatropha curcas] |
| 6 |
Hb_011628_010 |
0.0949137127 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 7 |
Hb_004705_060 |
0.1026680903 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 8 |
Hb_000813_020 |
0.1031760289 |
- |
- |
PREDICTED: metacaspase-3-like [Jatropha curcas] |
| 9 |
Hb_000333_140 |
0.1032081358 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 10 |
Hb_009367_070 |
0.1036902567 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 11 |
Hb_003427_070 |
0.1070015442 |
- |
- |
hypothetical protein POPTR_0067s00310g [Populus trichocarpa] |
| 12 |
Hb_003777_150 |
0.1085593685 |
- |
- |
PREDICTED: uncharacterized protein LOC105640938 isoform X1 [Jatropha curcas] |
| 13 |
Hb_016911_010 |
0.1122275632 |
- |
- |
hypothetical protein POPTR_0010s15010g [Populus trichocarpa] |
| 14 |
Hb_084304_010 |
0.1160179962 |
- |
- |
PREDICTED: probable auxin efflux carrier component 1c isoform X2 [Jatropha curcas] |
| 15 |
Hb_004732_070 |
0.1174462983 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_034713_010 |
0.1177913876 |
- |
- |
PREDICTED: uncharacterized protein LOC105133378 [Populus euphratica] |
| 17 |
Hb_051212_020 |
0.1204093968 |
- |
- |
PREDICTED: 24-methylenesterol C-methyltransferase 2-like [Jatropha curcas] |
| 18 |
Hb_043552_010 |
0.1251648654 |
- |
- |
PREDICTED: pectinesterase/pectinesterase inhibitor PPE8B isoform X1 [Jatropha curcas] |
| 19 |
Hb_002304_110 |
0.1260246981 |
- |
- |
hypothetical protein MIMGU_mgv1a023246mg [Erythranthe guttata] |
| 20 |
Hb_015292_020 |
0.1274898508 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |