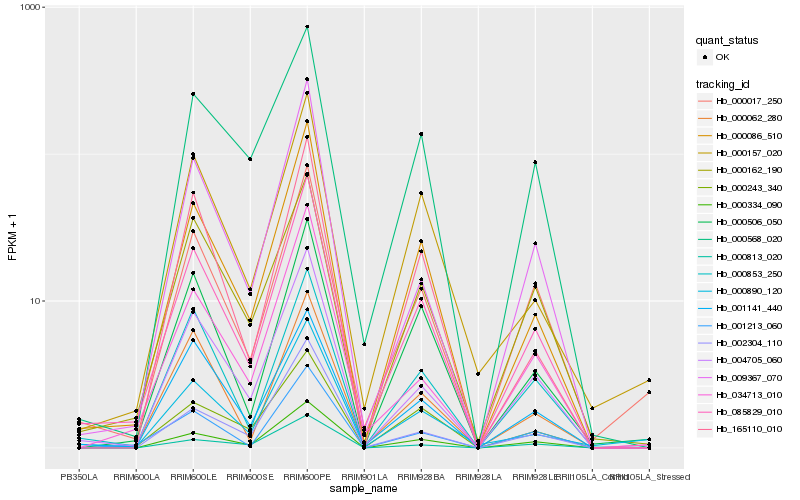
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_085829_010 |
0.0 |
- |
- |
PREDICTED: homeobox protein 2-like [Jatropha curcas] |
| 2 |
Hb_000086_510 |
0.0669896734 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_004705_060 |
0.0805706707 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 4 |
Hb_000334_090 |
0.0876880615 |
- |
- |
hypothetical protein JCGZ_16789 [Jatropha curcas] |
| 5 |
Hb_000017_250 |
0.0992996879 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA1B-like [Jatropha curcas] |
| 6 |
Hb_001213_060 |
0.1063082614 |
- |
- |
PREDICTED: uncharacterized protein LOC105646662 [Jatropha curcas] |
| 7 |
Hb_000853_250 |
0.1078164019 |
- |
- |
Cellulose synthase A catalytic subunit 3 [UDP-forming], putative [Ricinus communis] |
| 8 |
Hb_000243_340 |
0.1135001873 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 9 |
Hb_000506_050 |
0.1139185819 |
- |
- |
PREDICTED: thymidine kinase [Jatropha curcas] |
| 10 |
Hb_001141_440 |
0.1148389118 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 11 |
Hb_034713_010 |
0.1152340223 |
- |
- |
PREDICTED: uncharacterized protein LOC105133378 [Populus euphratica] |
| 12 |
Hb_009367_070 |
0.118458156 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 13 |
Hb_000157_020 |
0.1188178681 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000162_190 |
0.1188374048 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 15 |
Hb_000890_120 |
0.119863077 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000062_280 |
0.1230528107 |
- |
- |
plant mitotic spindle assembly checkpoint protein mad2, putative [Ricinus communis] |
| 17 |
Hb_002304_110 |
0.1246291533 |
- |
- |
hypothetical protein MIMGU_mgv1a023246mg [Erythranthe guttata] |
| 18 |
Hb_165110_010 |
0.1251036242 |
- |
- |
PREDICTED: kunitz-type elastase inhibitor BrEI-like [Jatropha curcas] |
| 19 |
Hb_000568_020 |
0.1253263146 |
- |
- |
PREDICTED: probable aquaporin PIP2-5 [Jatropha curcas] |
| 20 |
Hb_000813_020 |
0.1312733381 |
- |
- |
PREDICTED: metacaspase-3-like [Jatropha curcas] |