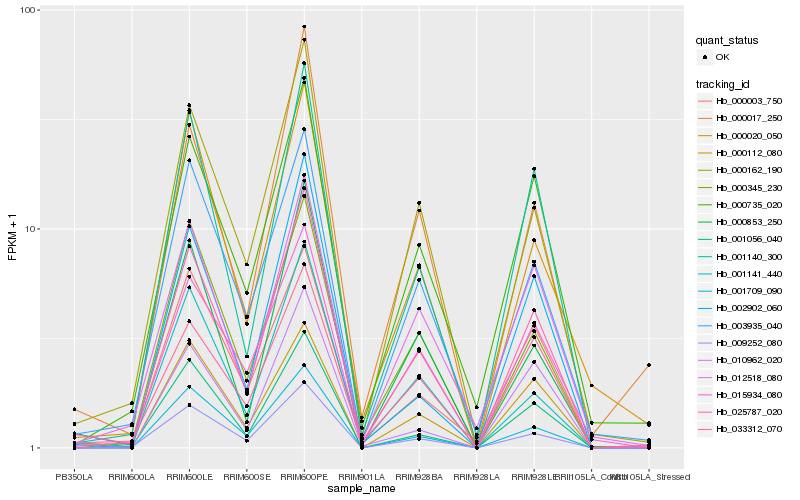
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025787_020 |
0.0 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 2 |
Hb_009252_080 |
0.0762403324 |
- |
- |
Polygalacturonase-1 non-catalytic subunit beta precursor, putative [Ricinus communis] |
| 3 |
Hb_001140_300 |
0.0765332035 |
- |
- |
PREDICTED: tubulin beta-1 chain [Cicer arietinum] |
| 4 |
Hb_000162_190 |
0.0791731698 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 5 |
Hb_001056_040 |
0.0881896182 |
- |
- |
hypothetical protein POPTR_0006s04450g [Populus trichocarpa] |
| 6 |
Hb_000345_230 |
0.0939912054 |
- |
- |
- |
| 7 |
Hb_001709_090 |
0.0964561072 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 8 |
Hb_000735_020 |
0.0970975521 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 9 |
Hb_003935_040 |
0.0998563973 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein SVP [Jatropha curcas] |
| 10 |
Hb_033312_070 |
0.1064112014 |
- |
- |
hypothetical protein POPTR_0017s02960g [Populus trichocarpa] |
| 11 |
Hb_000112_080 |
0.1067695216 |
- |
- |
PREDICTED: glutamate receptor 2.1-like [Populus euphratica] |
| 12 |
Hb_000853_250 |
0.1071302244 |
- |
- |
Cellulose synthase A catalytic subunit 3 [UDP-forming], putative [Ricinus communis] |
| 13 |
Hb_001141_440 |
0.1081372398 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 14 |
Hb_012518_080 |
0.1083088627 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like [Jatropha curcas] |
| 15 |
Hb_015934_080 |
0.1093643803 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 16 |
Hb_002902_060 |
0.1102331005 |
- |
- |
PREDICTED: uncharacterized protein LOC105639824 [Jatropha curcas] |
| 17 |
Hb_000017_250 |
0.1109715618 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA1B-like [Jatropha curcas] |
| 18 |
Hb_000020_050 |
0.1113118025 |
- |
- |
PREDICTED: expansin-A6 [Jatropha curcas] |
| 19 |
Hb_010962_020 |
0.1146172896 |
transcription factor |
TF Family: OFP |
hypothetical protein RCOM_0815310 [Ricinus communis] |
| 20 |
Hb_000003_750 |
0.1150068991 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Jatropha curcas] |