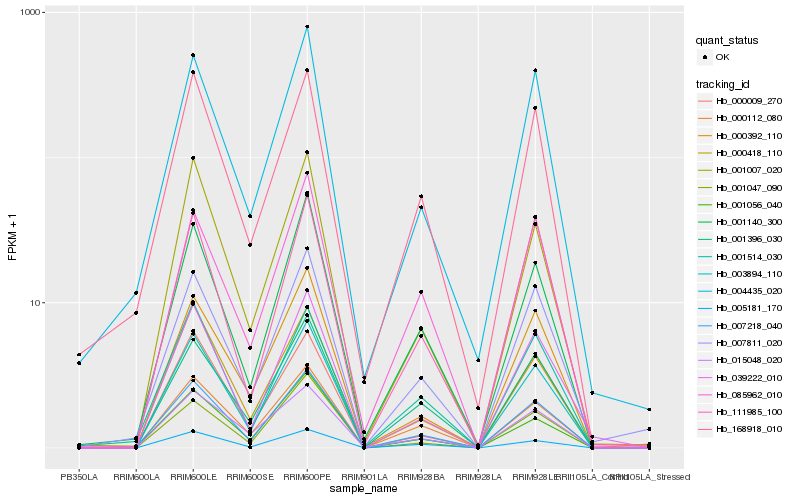
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007218_040 |
0.0 |
- |
- |
Leucine-rich repeat transmembrane protein kinase protein, putative [Theobroma cacao] |
| 2 |
Hb_001140_300 |
0.0736425336 |
- |
- |
PREDICTED: tubulin beta-1 chain [Cicer arietinum] |
| 3 |
Hb_001396_030 |
0.0744045794 |
- |
- |
hypothetical protein POPTR_0001s13520g [Populus trichocarpa] |
| 4 |
Hb_000112_080 |
0.0772908589 |
- |
- |
PREDICTED: glutamate receptor 2.1-like [Populus euphratica] |
| 5 |
Hb_168918_010 |
0.079277172 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 6 |
Hb_007811_020 |
0.0811083555 |
- |
- |
PREDICTED: expansin-A6 [Jatropha curcas] |
| 7 |
Hb_004435_020 |
0.0816585111 |
- |
- |
PREDICTED: tubulin alpha-2 chain [Jatropha curcas] |
| 8 |
Hb_000418_110 |
0.0846922025 |
- |
- |
PREDICTED: protein kinase PINOID [Jatropha curcas] |
| 9 |
Hb_039222_010 |
0.0862717235 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001514_030 |
0.0863885217 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 11 |
Hb_005181_170 |
0.086732184 |
- |
- |
PREDICTED: aldehyde dehydrogenase 22A1 [Jatropha curcas] |
| 12 |
Hb_001056_040 |
0.0883677767 |
- |
- |
hypothetical protein POPTR_0006s04450g [Populus trichocarpa] |
| 13 |
Hb_003894_110 |
0.091679216 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 14 |
Hb_001007_020 |
0.0935043871 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_015048_020 |
0.094111364 |
- |
- |
PREDICTED: myb-like protein D [Jatropha curcas] |
| 16 |
Hb_000392_110 |
0.0942959809 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 17 |
Hb_085962_010 |
0.0957323933 |
- |
- |
hypothetical protein POPTR_0017s11380g [Populus trichocarpa] |
| 18 |
Hb_000009_270 |
0.0993987183 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 19 |
Hb_001047_090 |
0.0995573181 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP16-like [Jatropha curcas] |
| 20 |
Hb_111985_100 |
0.1003863564 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |