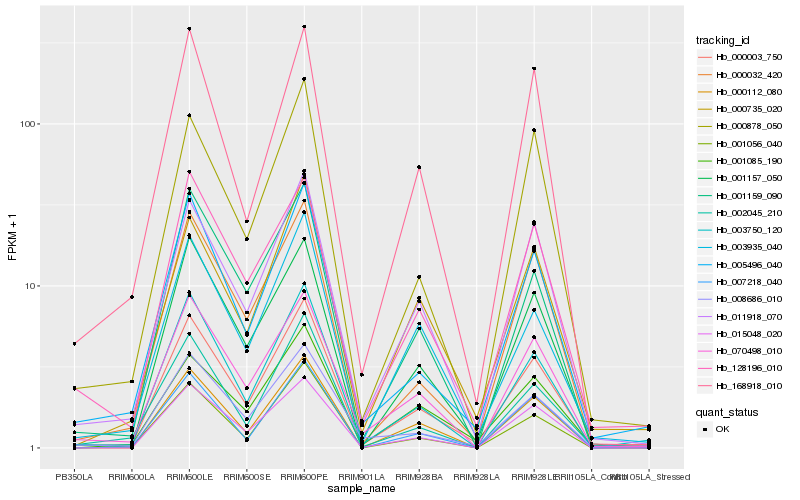
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_750 |
0.0 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Jatropha curcas] |
| 2 |
Hb_011918_070 |
0.0750456664 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 3 |
Hb_001157_050 |
0.0772291456 |
- |
- |
cation efflux protein/ zinc transporter, putative [Ricinus communis] |
| 4 |
Hb_003750_120 |
0.0840017514 |
- |
- |
PREDICTED: uncharacterized protein LOC105112992 [Populus euphratica] |
| 5 |
Hb_005496_040 |
0.0848872204 |
- |
- |
PREDICTED: uncharacterized protein LOC105630864 [Jatropha curcas] |
| 6 |
Hb_070498_010 |
0.0869016205 |
- |
- |
receptor kinase, putative [Ricinus communis] |
| 7 |
Hb_015048_020 |
0.0878934418 |
- |
- |
PREDICTED: myb-like protein D [Jatropha curcas] |
| 8 |
Hb_000032_420 |
0.0895708837 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 9 |
Hb_000112_080 |
0.0922257331 |
- |
- |
PREDICTED: glutamate receptor 2.1-like [Populus euphratica] |
| 10 |
Hb_001159_090 |
0.0969285301 |
- |
- |
PREDICTED: methylsterol monooxygenase 1-1-like [Jatropha curcas] |
| 11 |
Hb_003935_040 |
0.097507381 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein SVP [Jatropha curcas] |
| 12 |
Hb_008686_010 |
0.0986595972 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000878_050 |
0.0996090876 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 14 |
Hb_000735_020 |
0.1002541613 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 15 |
Hb_128196_010 |
0.1004813876 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_168918_010 |
0.1015972758 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 17 |
Hb_001056_040 |
0.1021727395 |
- |
- |
hypothetical protein POPTR_0006s04450g [Populus trichocarpa] |
| 18 |
Hb_002045_210 |
0.1031939847 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 19 |
Hb_007218_040 |
0.1036120915 |
- |
- |
Leucine-rich repeat transmembrane protein kinase protein, putative [Theobroma cacao] |
| 20 |
Hb_001085_190 |
0.1059826363 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |