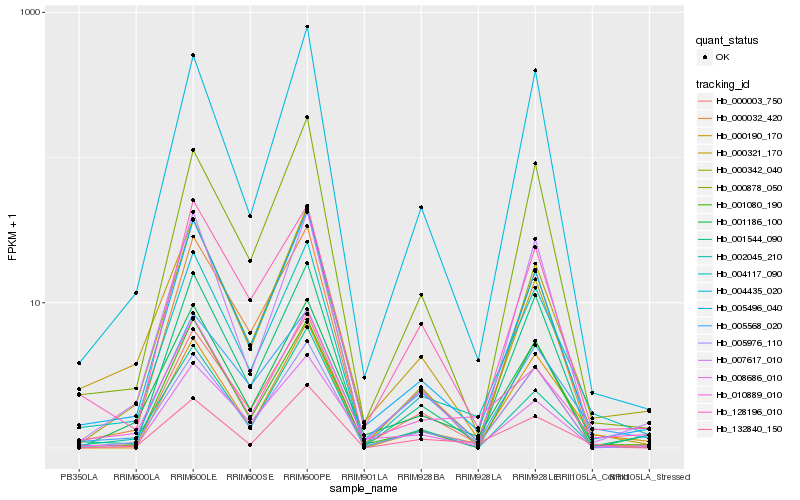
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004117_090 |
0.0 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 70 [Jatropha curcas] |
| 2 |
Hb_005496_040 |
0.0847737266 |
- |
- |
PREDICTED: uncharacterized protein LOC105630864 [Jatropha curcas] |
| 3 |
Hb_000342_040 |
0.0942545104 |
- |
- |
pollen specific protein sf21, putative [Ricinus communis] |
| 4 |
Hb_001544_090 |
0.1030742404 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 5 |
Hb_005976_110 |
0.1031917547 |
- |
- |
PREDICTED: uncharacterized protein LOC105639708 [Jatropha curcas] |
| 6 |
Hb_000190_170 |
0.1074466876 |
- |
- |
PREDICTED: chlorophyllase-1-like [Jatropha curcas] |
| 7 |
Hb_000032_420 |
0.1101625604 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 8 |
Hb_010889_010 |
0.1132587403 |
- |
- |
PREDICTED: myosin-12 [Jatropha curcas] |
| 9 |
Hb_001080_190 |
0.1176958687 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000003_750 |
0.117696669 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Jatropha curcas] |
| 11 |
Hb_000878_050 |
0.1182036038 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 12 |
Hb_004435_020 |
0.1187150297 |
- |
- |
PREDICTED: tubulin alpha-2 chain [Jatropha curcas] |
| 13 |
Hb_132840_150 |
0.1213064435 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 14 |
Hb_007617_010 |
0.1233416718 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001186_100 |
0.1236972805 |
- |
- |
PREDICTED: uncharacterized protein LOC105640806 isoform X1 [Jatropha curcas] |
| 16 |
Hb_128196_010 |
0.1241809849 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002045_210 |
0.1245919742 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 18 |
Hb_005568_020 |
0.1286831945 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 19 |
Hb_008686_010 |
0.1288719248 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000321_170 |
0.1293006537 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like [Jatropha curcas] |