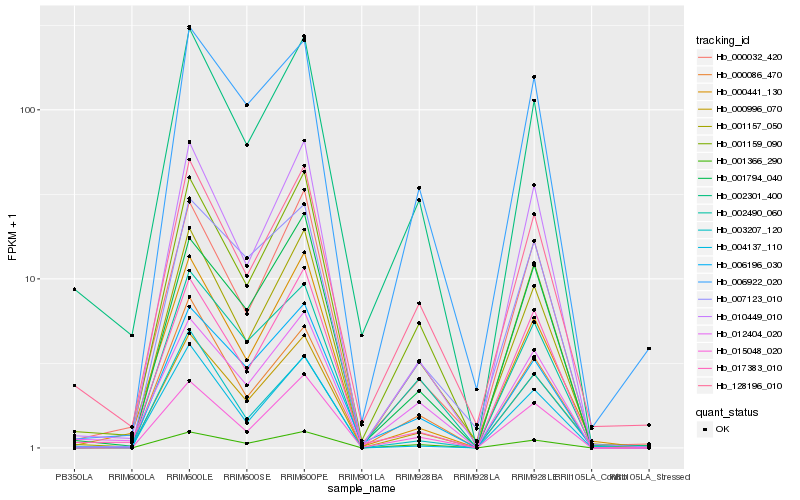
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000996_070 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_010449_010 |
0.0735403584 |
- |
- |
l-ascorbate oxidase, putative [Ricinus communis] |
| 3 |
Hb_000441_130 |
0.0765758293 |
- |
- |
PREDICTED: homeobox-leucine zipper protein HOX32 [Vitis vinifera] |
| 4 |
Hb_000032_420 |
0.0773763082 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 5 |
Hb_001157_050 |
0.0775351705 |
- |
- |
cation efflux protein/ zinc transporter, putative [Ricinus communis] |
| 6 |
Hb_006196_030 |
0.0870266965 |
- |
- |
PREDICTED: uncharacterized protein LOC105650679 [Jatropha curcas] |
| 7 |
Hb_001794_040 |
0.089771968 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 8 |
Hb_017383_010 |
0.0915411423 |
- |
- |
PREDICTED: uncharacterized protein LOC105628176 [Jatropha curcas] |
| 9 |
Hb_002301_400 |
0.0959487449 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 10 |
Hb_015048_020 |
0.0963259995 |
- |
- |
PREDICTED: myb-like protein D [Jatropha curcas] |
| 11 |
Hb_128196_010 |
0.1013045716 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_006922_020 |
0.1020187849 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |
| 13 |
Hb_007123_010 |
0.1025167077 |
- |
- |
PREDICTED: auxin efflux carrier component 3-like [Jatropha curcas] |
| 14 |
Hb_001159_090 |
0.1048634219 |
- |
- |
PREDICTED: methylsterol monooxygenase 1-1-like [Jatropha curcas] |
| 15 |
Hb_002490_060 |
0.1054093754 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 16 |
Hb_000086_470 |
0.1054752127 |
- |
- |
hypothetical protein JCGZ_17645 [Jatropha curcas] |
| 17 |
Hb_001366_290 |
0.1075105474 |
- |
- |
Alpha-expansin 1 precursor, putative [Ricinus communis] |
| 18 |
Hb_003207_120 |
0.109817308 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 19 |
Hb_012404_020 |
0.1105659135 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004137_110 |
0.1109014643 |
- |
- |
PREDICTED: probable auxin efflux carrier component 6 [Jatropha curcas] |