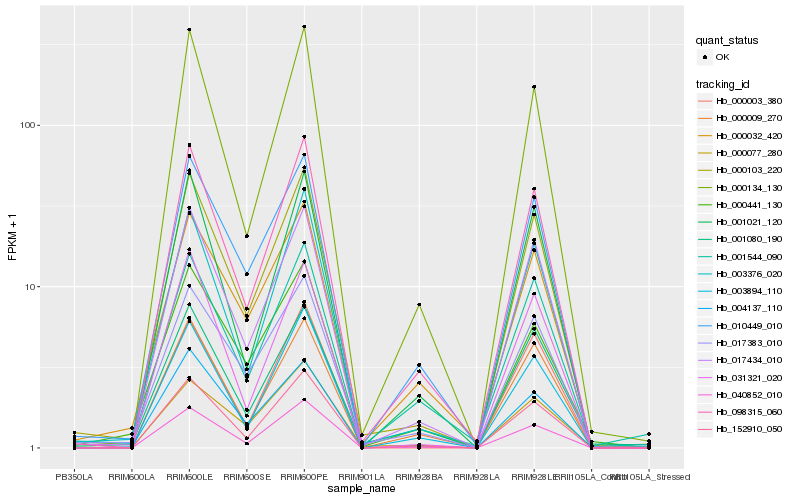
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000103_220 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632036 [Jatropha curcas] |
| 2 |
Hb_017434_010 |
0.0432029433 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 3 |
Hb_152910_050 |
0.0594022239 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_098315_060 |
0.0599179913 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 5 |
Hb_017383_010 |
0.0667880624 |
- |
- |
PREDICTED: uncharacterized protein LOC105628176 [Jatropha curcas] |
| 6 |
Hb_003376_020 |
0.0674763348 |
- |
- |
PREDICTED: interactor of constitutive active ROPs 2, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_040852_010 |
0.0704643952 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_010449_010 |
0.0726096479 |
- |
- |
l-ascorbate oxidase, putative [Ricinus communis] |
| 9 |
Hb_000134_130 |
0.0773865886 |
- |
- |
PREDICTED: uncharacterized protein LOC105628670 [Jatropha curcas] |
| 10 |
Hb_004137_110 |
0.083731217 |
- |
- |
PREDICTED: probable auxin efflux carrier component 6 [Jatropha curcas] |
| 11 |
Hb_003894_110 |
0.0880859273 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 12 |
Hb_000077_280 |
0.0904450024 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_031321_020 |
0.0912244349 |
- |
- |
hypothetical protein POPTR_0003s15520g [Populus trichocarpa] |
| 14 |
Hb_000032_420 |
0.0918501599 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 15 |
Hb_000009_270 |
0.0920158061 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 16 |
Hb_001021_120 |
0.0928272769 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-4 [Jatropha curcas] |
| 17 |
Hb_001080_190 |
0.0949409802 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000003_380 |
0.0961658268 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000441_130 |
0.0987056523 |
- |
- |
PREDICTED: homeobox-leucine zipper protein HOX32 [Vitis vinifera] |
| 20 |
Hb_001544_090 |
0.1011250435 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |