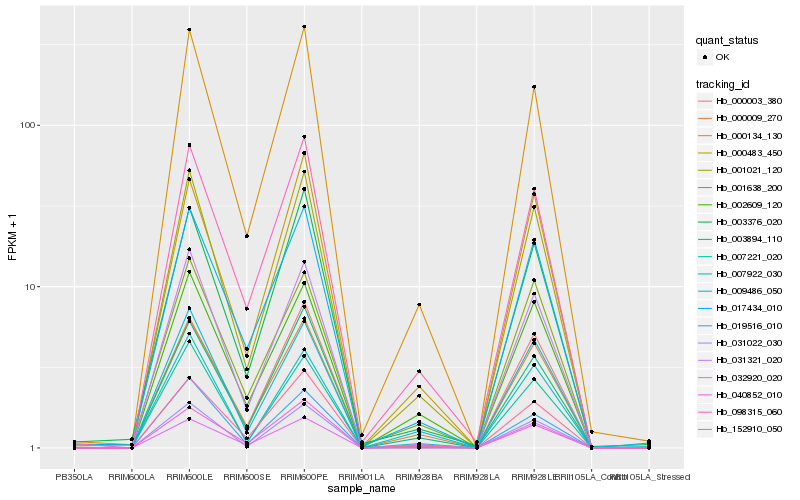
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001021_120 |
0.0 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-4 [Jatropha curcas] |
| 2 |
Hb_000009_270 |
0.0401361737 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 3 |
Hb_031321_020 |
0.0577439504 |
- |
- |
hypothetical protein POPTR_0003s15520g [Populus trichocarpa] |
| 4 |
Hb_000134_130 |
0.0595573245 |
- |
- |
PREDICTED: uncharacterized protein LOC105628670 [Jatropha curcas] |
| 5 |
Hb_007922_030 |
0.0606502509 |
- |
- |
PREDICTED: uncharacterized protein LOC105111213 [Populus euphratica] |
| 6 |
Hb_152910_050 |
0.0627283136 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_098315_060 |
0.0636879061 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 8 |
Hb_017434_010 |
0.0691598621 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 9 |
Hb_007221_020 |
0.0702985606 |
- |
- |
FAD-binding domain-containing family protein [Populus trichocarpa] |
| 10 |
Hb_003894_110 |
0.070569118 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 11 |
Hb_002609_120 |
0.0708731236 |
- |
- |
hypothetical protein POPTR_0001s25660g [Populus trichocarpa] |
| 12 |
Hb_031022_030 |
0.0713785227 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7-like [Jatropha curcas] |
| 13 |
Hb_009486_050 |
0.0717581497 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |
| 14 |
Hb_000483_450 |
0.0754305516 |
- |
- |
PREDICTED: anthranilate N-benzoyltransferase protein 1 [Jatropha curcas] |
| 15 |
Hb_019516_010 |
0.0794158971 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like isoform X2 [Populus euphratica] |
| 16 |
Hb_040852_010 |
0.0798421491 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_032920_020 |
0.0808930603 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 18 |
Hb_003376_020 |
0.0819032499 |
- |
- |
PREDICTED: interactor of constitutive active ROPs 2, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_001638_200 |
0.0843123071 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 43 [Jatropha curcas] |
| 20 |
Hb_000003_380 |
0.0881887643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |