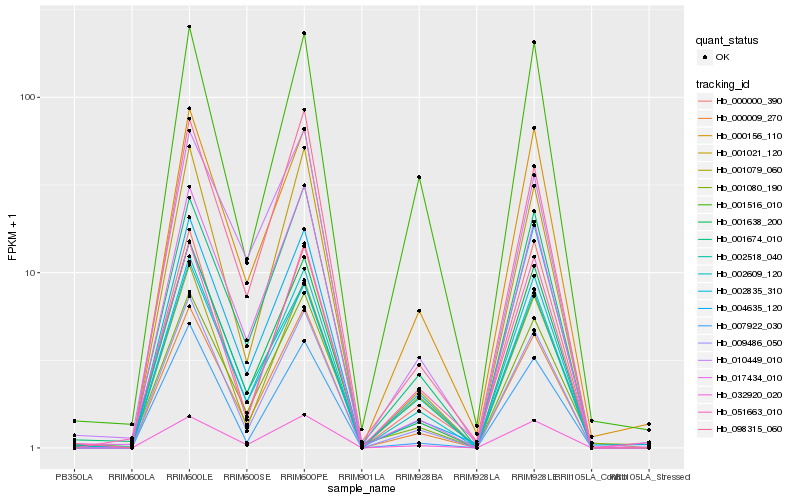
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002609_120 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s25660g [Populus trichocarpa] |
| 2 |
Hb_000009_270 |
0.0393009295 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 3 |
Hb_032920_020 |
0.0549028313 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_001638_200 |
0.0593502938 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 43 [Jatropha curcas] |
| 5 |
Hb_009486_050 |
0.0630126539 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |
| 6 |
Hb_000156_110 |
0.0659989564 |
- |
- |
Uncharacterized protein TCM_029490 [Theobroma cacao] |
| 7 |
Hb_001021_120 |
0.0708731236 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-4 [Jatropha curcas] |
| 8 |
Hb_001674_010 |
0.0769358193 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 9 |
Hb_002835_310 |
0.0783320196 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_001079_060 |
0.0797190252 |
- |
- |
PREDICTED: probable vacuolar amino acid transporter YPQ1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_017434_010 |
0.0802047519 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 12 |
Hb_001080_190 |
0.0853921349 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002518_040 |
0.0877283824 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 14 |
Hb_000000_390 |
0.0881754809 |
- |
- |
PREDICTED: uncharacterized protein LOC105633753 [Jatropha curcas] |
| 15 |
Hb_098315_060 |
0.088719028 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 16 |
Hb_007922_030 |
0.0905723337 |
- |
- |
PREDICTED: uncharacterized protein LOC105111213 [Populus euphratica] |
| 17 |
Hb_001516_010 |
0.0931848015 |
- |
- |
PREDICTED: guanylate kinase 2 [Jatropha curcas] |
| 18 |
Hb_004635_120 |
0.0937050963 |
- |
- |
PREDICTED: rho GTPase-activating protein 4-like [Jatropha curcas] |
| 19 |
Hb_051663_010 |
0.0945119298 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 20 |
Hb_010449_010 |
0.0962543511 |
- |
- |
l-ascorbate oxidase, putative [Ricinus communis] |