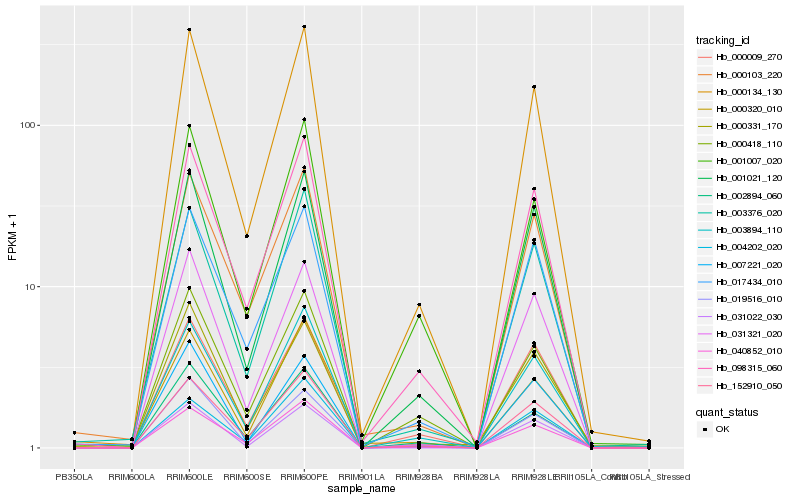
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000134_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628670 [Jatropha curcas] |
| 2 |
Hb_152910_050 |
0.047040894 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_098315_060 |
0.0481758634 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 4 |
Hb_040852_010 |
0.0489969941 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003894_110 |
0.0499196416 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 6 |
Hb_001021_120 |
0.0595573245 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-4 [Jatropha curcas] |
| 7 |
Hb_003376_020 |
0.0659169252 |
- |
- |
PREDICTED: interactor of constitutive active ROPs 2, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_019516_010 |
0.0700992446 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like isoform X2 [Populus euphratica] |
| 9 |
Hb_001007_020 |
0.0717323069 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_031321_020 |
0.0729625438 |
- |
- |
hypothetical protein POPTR_0003s15520g [Populus trichocarpa] |
| 11 |
Hb_000320_010 |
0.0753121859 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000103_220 |
0.0773865886 |
- |
- |
PREDICTED: uncharacterized protein LOC105632036 [Jatropha curcas] |
| 13 |
Hb_002894_060 |
0.0780176398 |
- |
- |
respiratory burst oxidase D [Manihot esculenta] |
| 14 |
Hb_017434_010 |
0.0782478744 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 15 |
Hb_000418_110 |
0.0784041882 |
- |
- |
PREDICTED: protein kinase PINOID [Jatropha curcas] |
| 16 |
Hb_000009_270 |
0.0784387617 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 17 |
Hb_031022_030 |
0.0865776726 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7-like [Jatropha curcas] |
| 18 |
Hb_007221_020 |
0.0869615093 |
- |
- |
FAD-binding domain-containing family protein [Populus trichocarpa] |
| 19 |
Hb_000331_170 |
0.0909148901 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At3g03770 [Jatropha curcas] |
| 20 |
Hb_004202_020 |
0.0959541697 |
- |
- |
conserved hypothetical protein [Ricinus communis] |