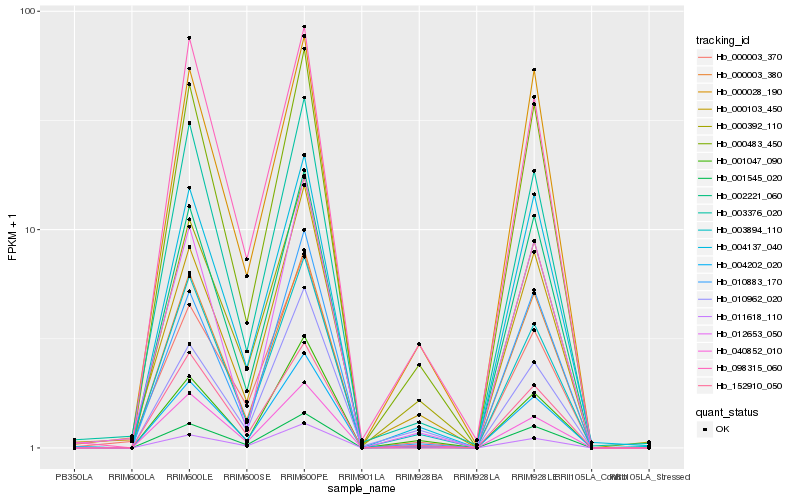
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004202_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000483_450 |
0.0487285887 |
- |
- |
PREDICTED: anthranilate N-benzoyltransferase protein 1 [Jatropha curcas] |
| 3 |
Hb_001047_090 |
0.0608111428 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP16-like [Jatropha curcas] |
| 4 |
Hb_040852_010 |
0.0609741819 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003894_110 |
0.0613776997 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 6 |
Hb_000392_110 |
0.0659902638 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 7 |
Hb_003376_020 |
0.0681988085 |
- |
- |
PREDICTED: interactor of constitutive active ROPs 2, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_152910_050 |
0.0684769302 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_010883_170 |
0.0725453662 |
- |
- |
PREDICTED: auxin transporter-like protein 3 [Jatropha curcas] |
| 10 |
Hb_011618_110 |
0.0767124224 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020-like [Citrus sinensis] |
| 11 |
Hb_000103_450 |
0.0778933761 |
- |
- |
PREDICTED: dentin sialophosphoprotein-like [Jatropha curcas] |
| 12 |
Hb_004137_040 |
0.0784372412 |
- |
- |
PREDICTED: serine carboxypeptidase-like 42 [Jatropha curcas] |
| 13 |
Hb_000003_370 |
0.0786827546 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 14 |
Hb_002221_060 |
0.0800108891 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 15 |
Hb_098315_060 |
0.0801901785 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 16 |
Hb_012653_050 |
0.0836341631 |
- |
- |
hypothetical protein RCOM_0460020 [Ricinus communis] |
| 17 |
Hb_000003_380 |
0.0840153588 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001545_020 |
0.0889576616 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X5 [Populus euphratica] |
| 19 |
Hb_000028_190 |
0.0892822887 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 20 |
Hb_010962_020 |
0.0898010355 |
transcription factor |
TF Family: OFP |
hypothetical protein RCOM_0815310 [Ricinus communis] |